Difference between revisions of "2010 Summer Project Week Quantification Of Lesion Diffusion Disruption"
From NAMIC Wiki
(Initial page) |
(Added lesion diffusion image) |
||
| Line 2: | Line 2: | ||
<gallery> | <gallery> | ||
Image:PW-MIT2010.png|[[2010_Summer_Project_Week#Projects|Projects List]] | Image:PW-MIT2010.png|[[2010_Summer_Project_Week#Projects|Projects List]] | ||
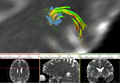
| − | Image: | + | Image:TractsLesionZoomOverlay.png|Tracts interrupted by lesions with FA values. |
| − | |||
</gallery> | </gallery> | ||
Revision as of 16:19, 21 June 2010
Home < 2010 Summer Project Week Quantification Of Lesion Diffusion Disruption
Key Investigators
- ABMIG: H. J. Bockholt
- MRN: Mark Scully
Objective
Quantify the effect lesions have on diffusion.
Approach, Plan
Our initial approach for quantifying diffusion interruption is to compile statistics on the distributions of FA at varying distances from lesion tissue. Assuming there are significant differences more advanced methods, such as using graph laplacians to estimate probable tracts will be investigated.
Progress
Using diffusion tools in slicer FA images were created in the same space as the lesion images. ITK code for summarizing statistics is almost complete.
Delivery Mechanism
This work will be delivered to the NA-MIC Kit as a Slicer3 commandline extension.