Difference between revisions of "2010 Winter Project Week WMLS"
| Line 17: | Line 17: | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
| − | We will continue developing and testing a Slicer module for the white matter lesion segmentation algorithm | + | We will continue developing and testing a Slicer module for the white matter lesion segmentation algorithm. |
</div> | </div> | ||
| Line 24: | Line 24: | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| − | We will improve the current version of WMLS Slicer module, especially the generalization performance of the training model and speed. | + | We will improve the current version of WMLS Slicer module, especially the generalization performance of the training model and speed. Moreover, we will do more extensive testing by obtaining more data within NA-MIC community. |
</div> | </div> | ||
Revision as of 19:03, 16 June 2010
Home < 2010 Winter Project Week WMLS
Key Investigators
- UNC: Minjeong Kim, Dinggang Shen
- GE : Xiaodong Tao, Jim Miller
Objective
We will continue developing and testing a Slicer module for the white matter lesion segmentation algorithm.
Approach, Plan
We will improve the current version of WMLS Slicer module, especially the generalization performance of the training model and speed. Moreover, we will do more extensive testing by obtaining more data within NA-MIC community.
Progress
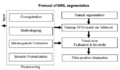
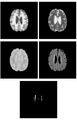
We have developed and tested a Slicer module for WML segmentation. Moreover, we have improved the speed of segmentation stage after training. We will provide the tutorial for this project meeting to present how to use our learning-based white matter lesion segmenation algorithm in Slicer 3. Subtasks implemented include: (1) a skull stripping algorithm working on T1 weighted images; (2) a fuzzy clustering algorithm for tissue segmentation; (3) a parametric model for gain field correction. All of these subtasks are implemented by using ITK. The training step uses AdaBoost and the segmenation step uses a support vector machine.
References
- Zhiqiang Lao, Dinggang Shen, Dengfeng Liu, Abbas F Jawad, Elias R Melhem, Lenore J Launer, Nick R Bryan, Christos Davatzikos, Computer-Assisted Segmentation of White Matter Lesions in 3D MR images, Using Pattern Recognition, Academic Radiology, 15(3):300-313, March 2008.