2011 Summer Project Week Integrate BRAINSCut into Slicer3
From NAMIC Wiki
Home < 2011 Summer Project Week Integrate BRAINSCut into Slicer3
Integrate BRAINSCut into Slicer3
Key Investigators
- University of Iowa: Eunyoung(Regina) Kim, Hans Johnson, Mark Scully
Objective
- Integration of BRAINSCut into Slicer3/Slicer4
- Integration of GMI feature images into BRAINSCut
- Test BRAINSCut
Approach, Plan
- Check dependencies of BRAINSCut and integrate into Slicer CLM.
- Process test data set to verify
- Submit caudate results to the CAUSE07 Challenge 'http://www.cause07.org/'
Progress
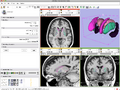
- BRAINSCut Incorporating with Slicer3 DONE
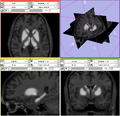
- Generating BRAINSCut model for subcortical structures: caudate, putamen, thalamus, and hippocampus. DONE
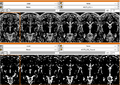
- Verifying generated model by comparing manual traces. DONE
- Process 230 data to generate hippocampus. DONE
References
- Powell, S., Magnotta, V., Johnson, H., and . . . , V. J., “Registration and machine learning-based automated segmentation of subcortical and cerebellar brain . . . ,” NeuroImage (Jan 2008)
- Eun Young Kim, Hans Johnson, "Multi-structure segmentation of multi-modal brain images using artificial neural networks", SPIE Proceedings Paper (Mar 2010)
Download Page
http://www.nitrc.org/projects/brainscut
Delivery Mechanism
This work will be delivered to the NAMIC Kit as a
- NITRIC distribution
- Slicer Module
- Built-in:
- Extension -- commandline:
- Extension -- loadable: