2011 Winter Project Week:RegistrationInPresenceOfAnatomicVariation
Key Investigators
- Kitware: Danielle Pace, Stephen Aylward
- UNC: Marc Niethammer
- SPL: Sandy Wells, Tina Kapur, Petter Risholm
Objective
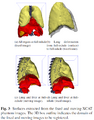
We are interested in registering images that contain anatomical variation between them, for example where organs slide against each other. For example, the brain may slide against the skull due to intraoperative brain shift, the heart slides relative to the lungs throughout the cardiac cycle, and respiration induces sliding of the lungs against the chest wall or abdominal muscles against each other. Not considering anatomical variation during image registration leads to errors that may impact clinical outcomes.
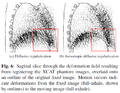
We have developed a regularization approach for deformable image registration that considers sliding motion and is based on anisotropic diffusion, and have shown using artificial and phantom images that it results in more plausible deformation fields compared to a typical diffusive (Gaussian smoothing) regularization.
Approach, Plan
During the project week, we will discuss and prototype potential improvements to the method, which may include:
- Organ border specification using the structure tensor (segmentation currently required)
- Incorporate more efficient diffusion calculation schemes / code profiling and optimization
- Representing normals as tensors rather than vectors, for improved performance at object corners
Progress
Very fruitful discussions on future work with Sandy Wells and Petter Risholm
Discussions and initial testing with Andriy Fedorov on applications to prostate interventions
Sliding organ registration profiling and optimizations:
- 18% faster in Debug mode
- 31% faster in Release mode
- (measured by registering two small 30x30x30 test images over 500 iterations)
Delivery Mechanism
This work will be delivered to the NA-MIC Kit as a:
- ITK Module
- Slicer Module
- Built-in
- Extension -- commandline YES
- Extension -- loadable
- Other YES
The sliding registration software is distributed as part of TubeTK, a new open-source toolkit providing software for registration, segmentation, analysis and quantification of images depicting tubular stuctures, such as vessels, bronchi and neurons. In particular, the sliding organ registration algorithm is incorporated into 3D Slicer as a CLI ("Anisotropic Diffusive Deformable Registration") that is provided with TubeTK (see the algorithm's page here).