2012 Winter Project Week: DTI Change Modeling
Key Partners
- Utah: Anuja Sharma, Guido Gerig
- Iowa: Hans Johnson (HD Project)
Objective
We have developed a new framework for spatiotemporal analysis of parameterized functions attributed by properties of 4D longitudinal image data. Our objective is to apply the framework to model patient-specific changes along white matter tracts using serial DTI data from subjects with Huntington's disease. As an extension, multiple subjects could be modeled and compared for assessment of normative trends.
Approach, Plan
- Co-register all available intra-subject, serial DTI volumes to create a consistent atlas tract geometry (and inter-subject if multiple subjects are to be compared eventually).
- Perform atlas-based tractography followed by back-mapping to assemble scalar diffusion parameters as functions of arc length along tracts of interest.
- Apply our methodology to quantify longitudinal DTI changes along tracts using these arc length parameterized diffusion functions.
Progress
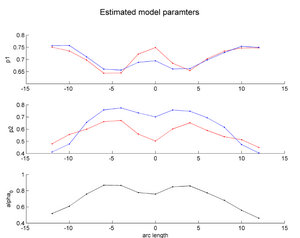
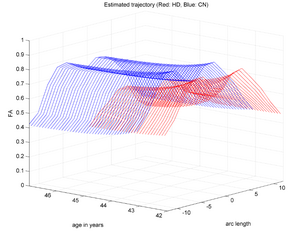
Co-registered a couple of HD and Control subjects. Performed tractography and back mapping to get discrete-time scalar diffusion curves for these subjects. Concept demonstration was done by quantifying patient specific longitudinal change trajectory for two subjects (one HD and one Control) simultaneously by estimating their individualized continuous spatiotemporal model and capturing the changes through model parameters. Details of the processing and preliminary results are discussed below.
White matter analysis for PREDICT-HD DBP
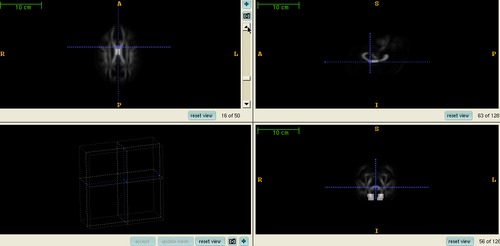
Registration and Tractography
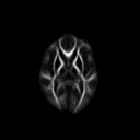
Tensor derived FA Atlas Slices from the mean atlas FA image computed by registering a couple of HD cases and Controls.
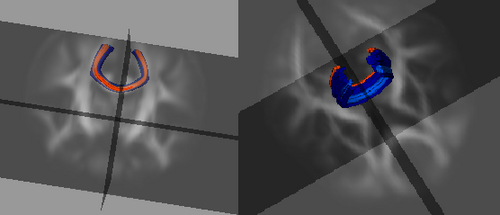
Tractography in atlas space for mid-corpus callosum tract Shown here are the source and target ROI placements along with the resulting fiber bundle representing the middle section of the corpus callosum white matter tract. This tract geometry is then mapped back to individual subjects to assemble subject specific diffusion information along the tract's length. The consistent across-subject tract definition provides a reference frame for comparison.
Longitudinal continuous spatiotemporal modeling of diffusion profiles for HD cases and Controls
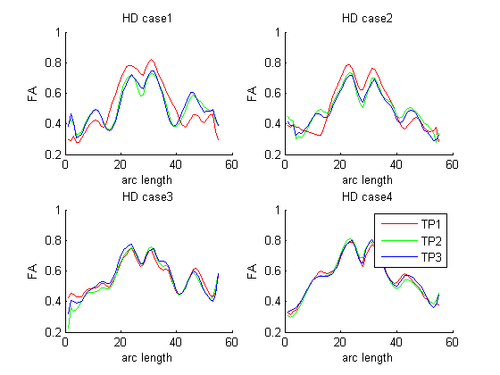
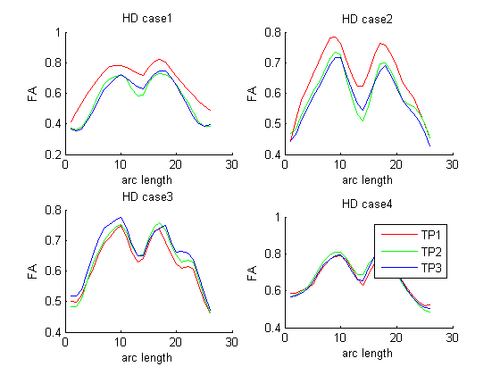
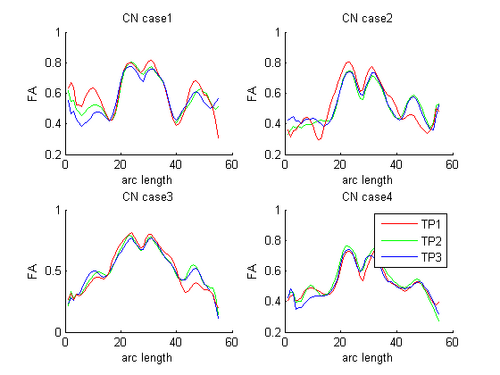
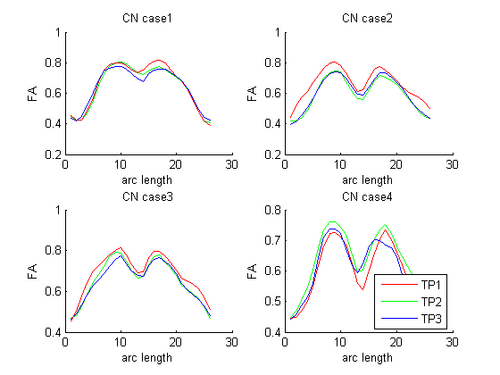
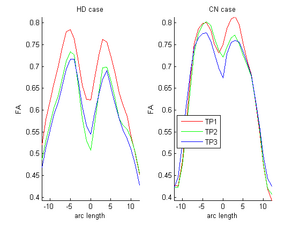
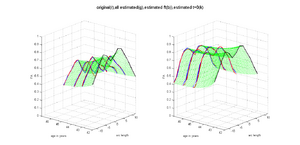
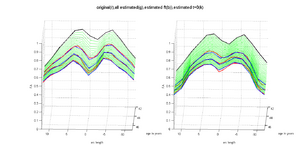
HD and Control subjects Shown below are the assembled along-tract diffusion profiles for a number of HD and Control subjects parametrized by arc length of the tract. The scalar diffusion measures (FA shown here) have been averaged across perpendicular cross sections along the tract's length. From what we see, the data is noisy as we do not see a consistent change pattern from timepoint 1 (TP1) to timepoint 3 (TP3) diffusion curves. At some places along the length, the FA values increase and then decrease while for some subjects, we see an overall decrease along length. Moreover, the ends of the tract are specially noisy. This could probably be improved by a more accurate registration and tractography procedure. However, since for this week our aim was a concept demonstration, we have worked on the middle of the diffusion curves (by clipping off the ends). Seen here are the plots for the complete tract as well as the middle portion for both HD and Control cases.
We demonstrate the feasibility of the method by applying the framework (Check link for details of the framework) to 2 subjects- one being a HD patient (10027) with a high burden factor (higher factor value (of factor 12) implying that the subjects is closer to the onset) and the other being a control subject (10004). Both have a similar baseline scan age (42 and 43 years respectively) as well as a similar time separation between follow up scans. This allows an intuitive comparison of the estimated growth trajectories.
This is a preliminary concept demonstration showing that the framework allows us to do patient-specific change modeling of serial HD data of a single patient given discrete-time diffusion curves.
References
- Sharma, A., Durrleman, S. , Gilmore, J.H. , Gerig, G. Longitudinal growth modeling of discrete-time functions with application to DTI tract evolution in early neurodevelopment. Proc. of 9th IEEE International Symposium on Biomedical Imaging (ISBI May'12), p.1397-1400.
- Casey B. Goodlett, P. Thomas Fletcher, John H. Gilmore, Guido Gerig. Group Analysis of DTI Fiber Tract Statistics with Application to Neurodevelopment. NeuroImage 45 (1) Supp. 1, 2009. p. S133-S142
- Details of the framework