2016 Winter Project Week/Projects/BatchImageAnalysis
From NAMIC Wiki
Home < 2016 Winter Project Week < Projects < BatchImageAnalysis
Key Investigators
- Kalli Retzepi (MGH)
- Yangming Ou (MGH)
- Matt Toews (ETS)
- Lilla Zollei (MGH)
- Lauren O'Donnell (BWH)
- Steve Pieper (BWH)
- Sandy Wells (BWH)
- Randy Gollub (MGH)
Project Description
| Objective | Approach and Plan | Progress and Next Steps |
|---|---|---|
|
|
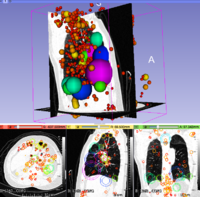
Algorithm
http://www.matthewtoews.com/fba/featExtract1.5.zip Result Baseline HIE classification rate: 73%, leave-one-out moderate vs normal.
|
Features Extracted in ADC MRI Volume