Difference between revisions of "2017 Winter Project Week/MeningiomaSegmentation"
From NAMIC Wiki
| Line 37: | Line 37: | ||
* Try Slicer in a Nipype workflow. | * Try Slicer in a Nipype workflow. | ||
* Apply manifold learning (a method of dimensionality reduction). | * Apply manifold learning (a method of dimensionality reduction). | ||
| + | * Get more data, and potentially train a neural network. | ||
|} | |} | ||
Revision as of 01:25, 13 January 2017
Home < 2017 Winter Project Week < MeningiomaSegmentationKey Investigators
- Satrajit Ghosh, MIT
- Omar Arnaout, Brigham and Women's Hospital
Project Description
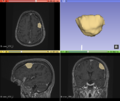
| Objective | Approach and Plan | Progress and Next Steps |
|---|---|---|
|
|
Progress
Next steps
|
Background and References
MR images of meningiomas that will be used in this project are available at OpenNeu.ro.