Difference between revisions of "Algorithm:MGH"
m (Fix MediaWiki table formatting issue discovered while converting to GitHub Flavored Markdown using pandoc (via https://github.com/outofcontrol/mediawiki-to-gfm)) Tag: 2017 source edit |
|||
| (31 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| − | Back to [[Algorithm:Main|NA-MIC Algorithms]] | + | Back to [[Algorithm:Main|NA-MIC Algorithms]] |
| − | + | __NOTOC__ | |
| − | = Overview of MGH Algorithms = | + | = Overview of MGH Algorithms (PI: Bruce Fischl) = |
A brief overview of the MGH's algorithms goes here. This should not be much longer than a paragraph. Remember that people visiting your site want to be able to understand very quickly what you're all about and then they want to jump into your site's projects. The projects below are organized into a two column table: the left column is for representative images and the right column is for project overviews. The number of rows corresponds to the number of projects at your site. Put the most interesting and relevant projects at the top of the table. You do not need to organize the table according to subject matter (i.e. do not group all segmentation projects together and all DWI projects together). | A brief overview of the MGH's algorithms goes here. This should not be much longer than a paragraph. Remember that people visiting your site want to be able to understand very quickly what you're all about and then they want to jump into your site's projects. The projects below are organized into a two column table: the left column is for representative images and the right column is for project overviews. The number of rows corresponds to the number of projects at your site. Put the most interesting and relevant projects at the top of the table. You do not need to organize the table according to subject matter (i.e. do not group all segmentation projects together and all DWI projects together). | ||
| Line 7: | Line 7: | ||
= MGH Projects = | = MGH Projects = | ||
| − | {| | + | {| cellpadding="10" style="text-align:left;" |
| − | + | || [[Image:Salatneuroimage09fig1.png|250px]] | |
| − | | | + | || |
| + | |||
| + | == [[Projects:ShapeBasedLevelSetSegmentation|Shape Based Level Segmentation]] == | ||
| + | |||
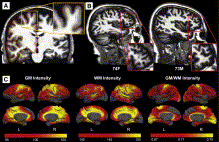
| + | This class of algorithms explicitly manipulates the representation of the object boundary to fit the strong gradients in the image, indicative of the object outline. Bias in the boundary evolution towards the likely shapes improves the robustness of the segmentation results when the intensity information alone is insufficient for boundary detection. [[Projects:ShapeBasedLevelSetSegmentation|More...]] | ||
| + | |||
| + | <font color="red">'''New: '''</font> Salat D.H., Lee S.Y., van der Kouwe A.J., Greve D.N., Fischl B., Rosas H.D. | ||
| + | Age-associated alterations in cortical gray and white matter signal intensity and gray to white matter contrast. Neuroimage. 2009 Oct 15;48(1):21-8. | ||
| + | |||
| + | |||
| + | |- | ||
| + | |||
| + | || [[Image:lh.pm14686.BA2.gif|250px]] | ||
| + | || | ||
| + | |||
| + | == [[Projects:LearningRegistrationCostFunctions| Learning Task-Optimal Registration Cost Functions]] == | ||
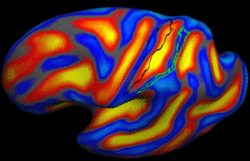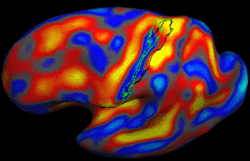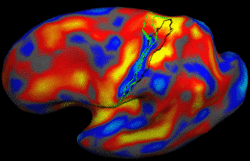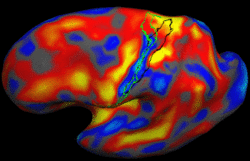
| − | + | We present a framework for learning the parameters of registration cost functions. The parameters of the registration cost function -- for example, the tradeoff between the image similarity and regularization terms -- are typically determined manually through inspection of the image alignment and then fixed for all applications. We propose a principled approach to learn these parameters with respect to particular applications. [[Projects:LearningRegistrationCostFunctions|More...]] | |
| − | + | <font color="red">'''New: '''</font> B.T.T. Yeo, M. Sabuncu, P. Golland, B. Fischl. Task-Optimal Registration Cost Functions. To appear in Proceedings of the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), 2009. | |
| − | + | |- | |
| + | |||
| + | | | [[Image:qdec.jpg|200px]] | ||
| + | | | | ||
| + | |||
| + | == [[Projects:QDEC|QDEC: An easy to use GUI for group morphometry studies]] == | ||
| + | |||
| + | Qdec is a application included in the Freesurfer software package intended to aid researchers in performing inter-subject / group averaging and inference on the morphometry data (cortical surface and volume) produced by the Freesurfer processing stream. The functionality in Qdec is also available as a processing module within Slicer3, and XNAT. [[Projects:QDEC|More...]] | ||
See: [http://surfer.nmr.mgh.harvard.edu/fswiki/Qdec Qdec user page] | See: [http://surfer.nmr.mgh.harvard.edu/fswiki/Qdec Qdec user page] | ||
| Line 21: | Line 43: | ||
|- | |- | ||
| − | | | [[Image: | + | | | [[Image:nrrd256.jpg|200px]] |
| − | | | | + | | | |
| + | == [[Projects:NRDDFreesurfer|Adding NRRD I/O to Freesurfer]] == | ||
| + | Our objective is to open a NRRD volume in FreeSurfer, and convert an MGH volume to a NRRD volume with Freesurfer. This project allows the seemless exchange of diffusion-based volumetric data between Slicer and the FreeSurfer analysis stream, including tensors, eigendirections, as well as raw muli-direction diffusion data. [[Projects:NRDDFreesurfer|More...]] | ||
| + | |- | ||
| + | | | [[Image:overcomplete_vs_biorthogonal_wavelets.jpg|200px]] | ||
| + | | | | ||
| − | == [[ | + | == [[Projects:CorticalSurfaceShapeAnalysisUsingSphericalWavelets|Spherical Wavelets]] == |
| + | Cortical Surface Shape Analysis Based on Spherical Wavelets. We introduce the use of over-complete spherical wavelets for shape analysis of 2D closed surfaces. Bi-orthogonal spherical wavelets have been proved to be powerful tools in the segmentation and shape analysis of 2D closed surfaces, but unfortunately they suffer from aliasing problems and are therefore not invariant to rotation of the underlying surface parameterization. In this paper, we demonstrate the theoretical advantage of over-complete wavelets over bi-orthogonal wavelets and illustrate their utility on both synthetic and real data. In particular, we show that the over-complete spherical wavelet transform enjoys significant advantages for the analysis of cortical folding development in a newborn dataset. [[Projects:CorticalSurfaceShapeAnalysisUsingSphericalWavelets|More...]] | ||
| − | + | |- | |
| − | |||
| − | == [[ | + | | | [[Image:separating_loops.jpg|200px]] |
| − | Cortical | + | | | |
| + | |||
| + | == [[Projects:TopologyCorrectionNonSeparatingLoops|Topology Correction]] == | ||
| + | |||
| + | Geometrically-Accurate Topology-Correction of Cortical Surfaces using Non-Separating Loops. We propose a technique to accurately correct the spherical topology of cortical surfaces. Specifically,we construct a mapping from the original surface onto the sphere to detect topological defects as minimal nonhomeomorphic regions. The topology of each defect is then corrected by opening and sealing the surface along a set of nonseparating loops that are selected in a Bayesian framework. The proposed method is a wholly self-contained topology correction algorithm, which determines geometrically accurate, topologically correct solutions based on the magnetic resonance imaging (MRI) intensity profile and the expected local curvature. Applied to real data, our method provides topological corrections similar to those made by a trained operator. [[Projects:TopologyCorrectionNonSeparatingLoops|More...]] | ||
|- | |- | ||
| − | | | [[Image: | + | | | [[Image:vxl.gif|200px]] |
| | | | | | ||
| − | == [[ | + | == [[Projects:FreeSurferNumericalRecipiesReplacement|Numerical Recipies Replacement]] == |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | Our objective is to replace algorithms using the proprietary Numerical Recipes for C source base in FreeSurfer in the efforts to open-source FreeSurfer. This project has been completed through the use of the open source packages VXL (VNL) and Cephes. This includes the complete replacement of all Numerical Recipes in C code, and the implementation of a battery of unit tests for each replaced function. Currently the open source release is at a beta stage, and 25 beta releases of the source have been made. We anticipate a complete open source release in first quarter 2008. [[Projects:FreeSurferNumericalRecipiesReplacement|More...]] | |
|- | |- | ||
| − | | | [[Image: | + | | | [[Image:histo_matching.jpg|200px]] |
| | | | | | ||
| − | == [[ | + | == [[Projects:AutomaticFullBrainSegmentation|Atlas Renormalization for Improved Brain MR Image Segmentation across Scanner Platforms]] == |
| − | + | Atlas-based approaches have demonstrated the ability to automatically identify detailed brain structures from 3-D magnetic resonance (MR) brain images. Unfortunately, the accuracy of this type of method often degrades when processing data acquired on a different scanner platform or pulse sequence than the data used for the atlas training. In this paper, we improve the performance of an atlas-based whole brain segmentation method by introducing an intensity renormalization procedure that automatically adjusts the prior atlas intensity model to new input data. Validation using manually labeled test datasets has shown that the new procedure improves the segmentation accuracy (as measured by the Dice coefficient) by 10% or more for several structures including hippocampus, amygdala, caudate, and pallidum. The results verify that this new procedure reduces the sensitivity of the whole brain segmentation method to changes in scanner platforms and improves its accuracy and robustness, which can thus facilitate multicenter or multisite neuroanatomical imaging studies. [[Projects:AutomaticFullBrainSegmentation|More...]] | |
| − | |||
| − | |||
|- | |- | ||
| − | | | [[Image: | + | | | [[Image:GT-SulciOutlining1.jpg|200px]] |
| − | | | | + | | | |
| − | == [[ | + | == [[Projects:SulciOutlining|Automatic Outlining of sulci on the brain surface]] == |
| − | + | We present a method to automatically extract certain key features on a surface. We apply this technique to outline sulci on the cortical surface of a brain. [[Projects:SulciOutlining|More...]] | |
| − | + | |} | |
Latest revision as of 06:04, 11 April 2023
Home < Algorithm:MGHBack to NA-MIC Algorithms
Overview of MGH Algorithms (PI: Bruce Fischl)
A brief overview of the MGH's algorithms goes here. This should not be much longer than a paragraph. Remember that people visiting your site want to be able to understand very quickly what you're all about and then they want to jump into your site's projects. The projects below are organized into a two column table: the left column is for representative images and the right column is for project overviews. The number of rows corresponds to the number of projects at your site. Put the most interesting and relevant projects at the top of the table. You do not need to organize the table according to subject matter (i.e. do not group all segmentation projects together and all DWI projects together).
MGH Projects
|
Shape Based Level SegmentationThis class of algorithms explicitly manipulates the representation of the object boundary to fit the strong gradients in the image, indicative of the object outline. Bias in the boundary evolution towards the likely shapes improves the robustness of the segmentation results when the intensity information alone is insufficient for boundary detection. More... New: Salat D.H., Lee S.Y., van der Kouwe A.J., Greve D.N., Fischl B., Rosas H.D. Age-associated alterations in cortical gray and white matter signal intensity and gray to white matter contrast. Neuroimage. 2009 Oct 15;48(1):21-8.
|
|
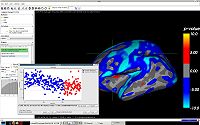
Learning Task-Optimal Registration Cost FunctionsWe present a framework for learning the parameters of registration cost functions. The parameters of the registration cost function -- for example, the tradeoff between the image similarity and regularization terms -- are typically determined manually through inspection of the image alignment and then fixed for all applications. We propose a principled approach to learn these parameters with respect to particular applications. More... New: B.T.T. Yeo, M. Sabuncu, P. Golland, B. Fischl. Task-Optimal Registration Cost Functions. To appear in Proceedings of the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), 2009. |
|
QDEC: An easy to use GUI for group morphometry studiesQdec is a application included in the Freesurfer software package intended to aid researchers in performing inter-subject / group averaging and inference on the morphometry data (cortical surface and volume) produced by the Freesurfer processing stream. The functionality in Qdec is also available as a processing module within Slicer3, and XNAT. More... See: Qdec user page |

|
Adding NRRD I/O to FreesurferOur objective is to open a NRRD volume in FreeSurfer, and convert an MGH volume to a NRRD volume with Freesurfer. This project allows the seemless exchange of diffusion-based volumetric data between Slicer and the FreeSurfer analysis stream, including tensors, eigendirections, as well as raw muli-direction diffusion data. More... |

|
Spherical WaveletsCortical Surface Shape Analysis Based on Spherical Wavelets. We introduce the use of over-complete spherical wavelets for shape analysis of 2D closed surfaces. Bi-orthogonal spherical wavelets have been proved to be powerful tools in the segmentation and shape analysis of 2D closed surfaces, but unfortunately they suffer from aliasing problems and are therefore not invariant to rotation of the underlying surface parameterization. In this paper, we demonstrate the theoretical advantage of over-complete wavelets over bi-orthogonal wavelets and illustrate their utility on both synthetic and real data. In particular, we show that the over-complete spherical wavelet transform enjoys significant advantages for the analysis of cortical folding development in a newborn dataset. More... |
Topology CorrectionGeometrically-Accurate Topology-Correction of Cortical Surfaces using Non-Separating Loops. We propose a technique to accurately correct the spherical topology of cortical surfaces. Specifically,we construct a mapping from the original surface onto the sphere to detect topological defects as minimal nonhomeomorphic regions. The topology of each defect is then corrected by opening and sealing the surface along a set of nonseparating loops that are selected in a Bayesian framework. The proposed method is a wholly self-contained topology correction algorithm, which determines geometrically accurate, topologically correct solutions based on the magnetic resonance imaging (MRI) intensity profile and the expected local curvature. Applied to real data, our method provides topological corrections similar to those made by a trained operator. More... | |

|
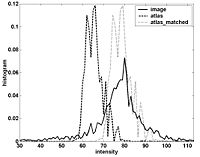
Numerical Recipies ReplacementOur objective is to replace algorithms using the proprietary Numerical Recipes for C source base in FreeSurfer in the efforts to open-source FreeSurfer. This project has been completed through the use of the open source packages VXL (VNL) and Cephes. This includes the complete replacement of all Numerical Recipes in C code, and the implementation of a battery of unit tests for each replaced function. Currently the open source release is at a beta stage, and 25 beta releases of the source have been made. We anticipate a complete open source release in first quarter 2008. More... |
|
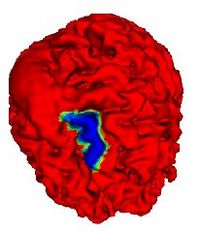
Atlas Renormalization for Improved Brain MR Image Segmentation across Scanner PlatformsAtlas-based approaches have demonstrated the ability to automatically identify detailed brain structures from 3-D magnetic resonance (MR) brain images. Unfortunately, the accuracy of this type of method often degrades when processing data acquired on a different scanner platform or pulse sequence than the data used for the atlas training. In this paper, we improve the performance of an atlas-based whole brain segmentation method by introducing an intensity renormalization procedure that automatically adjusts the prior atlas intensity model to new input data. Validation using manually labeled test datasets has shown that the new procedure improves the segmentation accuracy (as measured by the Dice coefficient) by 10% or more for several structures including hippocampus, amygdala, caudate, and pallidum. The results verify that this new procedure reduces the sensitivity of the whole brain segmentation method to changes in scanner platforms and improves its accuracy and robustness, which can thus facilitate multicenter or multisite neuroanatomical imaging studies. More... |
|
Automatic Outlining of sulci on the brain surfaceWe present a method to automatically extract certain key features on a surface. We apply this technique to outline sulci on the cortical surface of a brain. More... |