Difference between revisions of "DBP2:MIND:RoadmapProject"
Hjbockholt (talk | contribs) (New page: {| |thumb|320px|Return to [[2008_Summer_Project_Week|Project Week Main Page ]] |[[Image:genuFAp.jpg|thumb|320px|Scatter plot of the original FA data through ...) |
Hjbockholt (talk | contribs) |
||
| (25 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
{| | {| | ||
|[[Image:ProjectWeek-2008.png|thumb|320px|Return to [[2008_Summer_Project_Week|Project Week Main Page]] ]] | |[[Image:ProjectWeek-2008.png|thumb|320px|Return to [[2008_Summer_Project_Week|Project Week Main Page]] ]] | ||
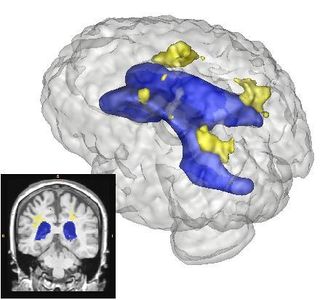
| − | |[[Image: | + | |[[Image:Lupus Demo001.png|thumb|320px|Example Results of Lesion Classification]] |
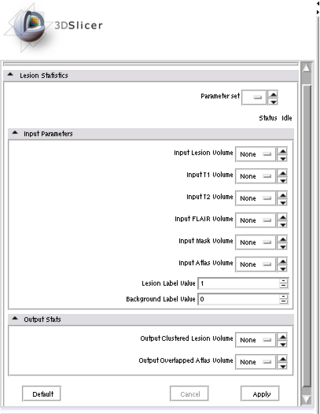
| − | |[[Image: | + | |[[Image:Lesion module.png|thumb|320px|Current Slicer3 Module]] |
|} | |} | ||
| Line 8: | Line 8: | ||
__NOTOC__ | __NOTOC__ | ||
| − | + | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
===Key Investigators=== | ===Key Investigators=== | ||
| − | * | + | *H. Jeremy Bockholt |
| − | * | + | *Mark Scully |
| − | + | *Ross Whitaker | |
| + | *Steve Pieper | ||
| + | *Vincent Magnotta | ||
| + | *Kilian Pohl | ||
| + | *Brad Davis | ||
| + | *Marcel Prastawa | ||
| + | *Guido Gerig | ||
| + | *Sonja Pujol | ||
<div style="margin: 20px;"> | <div style="margin: 20px;"> | ||
| Line 25: | Line 27: | ||
<h1>Objective</h1> | <h1>Objective</h1> | ||
| − | + | Finish the roadmap module, design and implement a testing strategy and work on the end-to-end tutorial for white matter lesion classification | |
| − | |||
| − | |||
</div> | </div> | ||
| Line 33: | Line 33: | ||
<h1>Approach, Plan</h1> | <h1>Approach, Plan</h1> | ||
| + | The Slicer3 module will allow co-registration of user-selected T1, T2, and FLAIR images. The module will automatically label all brain voxels into either gray, white, csf, or lesion as tissues types. Lesions will be automatically clustered such that the anatomical location and volume can be summarized. | ||
| + | |||
| + | Review and summarize results of each of classification approaches thus far. | ||
| + | |||
| + | Sketch out methods paper and clinical application paper. | ||
| + | |||
| + | Finalize content to be presented for SFN 2008 poster/presentation. | ||
| + | |||
| + | Work with Sonja on timeline and process for creating tutorial and process for release of tool and data (SFN 2008)? | ||
| + | |||
| + | Answer question, where do roadmap projects that share data, share it from, publication database, BIRN project, XNAT Central, other service, provide own? | ||
| − | |||
| − | |||
</div> | </div> | ||
| Line 42: | Line 51: | ||
<h1>Progress</h1> | <h1>Progress</h1> | ||
| + | Plan is to release end-to-end tutorial at the 2008 Society for Neuroscience Annual meeting. | ||
| + | |||
| + | Deliverables for this week | ||
| + | #Laid out the logic and components of the Joint Histogram matching filter for Intensity Standardization based on treating joint histograms as images and registering them to construct a deformation field. | ||
| + | #Integrated visualization of ROIs that lesions impinge upon using the volume renderer and met with Wendy on integrating the output with the Query Atlas. | ||
| + | #interested in working with the unbiased atlas work for lesion location (talk with sylvain) | ||
| + | #Met with Killian on tuning the parametization of EM-Segment to improve our lesion classification. | ||
| + | |||
| + | In terms of fine tuning lesion classification: | ||
| + | #We worked with Kilian on parameterization of EM-Segment | ||
| + | #We worked with Marcel on using his method for lesion classification | ||
| + | #We worked with Vince Magnotta on his method for lesion classification | ||
| + | |||
| + | Future Work: | ||
| − | |||
</div> | </div> | ||
| Line 50: | Line 72: | ||
</div> | </div> | ||
| − | |||
===References=== | ===References=== | ||
| − | |||
| − | |||
| − | |||
| − | |||
Latest revision as of 14:07, 27 June 2008
Home < DBP2:MIND:RoadmapProject Return to Project Week Main Page |
Key Investigators
- H. Jeremy Bockholt
- Mark Scully
- Ross Whitaker
- Steve Pieper
- Vincent Magnotta
- Kilian Pohl
- Brad Davis
- Marcel Prastawa
- Guido Gerig
- Sonja Pujol
Objective
Finish the roadmap module, design and implement a testing strategy and work on the end-to-end tutorial for white matter lesion classification
Approach, Plan
The Slicer3 module will allow co-registration of user-selected T1, T2, and FLAIR images. The module will automatically label all brain voxels into either gray, white, csf, or lesion as tissues types. Lesions will be automatically clustered such that the anatomical location and volume can be summarized.
Review and summarize results of each of classification approaches thus far.
Sketch out methods paper and clinical application paper.
Finalize content to be presented for SFN 2008 poster/presentation.
Work with Sonja on timeline and process for creating tutorial and process for release of tool and data (SFN 2008)?
Answer question, where do roadmap projects that share data, share it from, publication database, BIRN project, XNAT Central, other service, provide own?
Progress
Plan is to release end-to-end tutorial at the 2008 Society for Neuroscience Annual meeting.
Deliverables for this week
- Laid out the logic and components of the Joint Histogram matching filter for Intensity Standardization based on treating joint histograms as images and registering them to construct a deformation field.
- Integrated visualization of ROIs that lesions impinge upon using the volume renderer and met with Wendy on integrating the output with the Query Atlas.
- interested in working with the unbiased atlas work for lesion location (talk with sylvain)
- Met with Killian on tuning the parametization of EM-Segment to improve our lesion classification.
In terms of fine tuning lesion classification:
- We worked with Kilian on parameterization of EM-Segment
- We worked with Marcel on using his method for lesion classification
- We worked with Vince Magnotta on his method for lesion classification
Future Work: