Difference between revisions of "Events:August2008XNATDesktop"
m (Text replacement - "http://www.slicer.org/slicerWiki/index.php/" to "https://www.slicer.org/wiki/") |
|||
| (11 intermediate revisions by 2 users not shown) | |||
| Line 17: | Line 17: | ||
== Plan for supporting initial use case == | == Plan for supporting initial use case == | ||
| + | |||
| + | * Demonstrate this functionality at BIRN meeting in October | ||
| + | * First tests to be done by first week in September | ||
| + | * This plan to be presented to the NA-MIC Engineering tcon on Thursday August 28 at 3pm eastern. | ||
| + | |||
[[image:NeurosurgeryUsecase.png ]] | [[image:NeurosurgeryUsecase.png ]] | ||
| Line 28: | Line 33: | ||
Query for all subjects on system... | Query for all subjects on system... | ||
| − | '''send:''' curl | + | '''send:''' curl $srv/search??subject |
'''response:''' xml for all subjects in xnd.bwh.harvard.edu | '''response:''' xml for all subjects in xnd.bwh.harvard.edu | ||
Choose a subject, query for available data... | Choose a subject, query for available data... | ||
| − | '''send:''' curl | + | '''send:''' curl $srv/search?subject=$subj |
'''response:''' xml containing URIs of all resources for the individual subject $subj | '''response:''' xml containing URIs of all resources for the individual subject $subj | ||
Slicer parses XML and presents list of all resources; researcher2 queries for for only DICOM data for a particular subject... | Slicer parses XML and presents list of all resources; researcher2 queries for for only DICOM data for a particular subject... | ||
| − | '''send:''' curl | + | '''send:''' curl $srv/search?subject=$subj&file_type=DICOM |
'''response:''' xml containing URIs of all DICOM resources for subject $subj | '''response:''' xml containing URIs of all DICOM resources for subject $subj | ||
Slicer parses XML and presents list of URIs, researcher2 requests to download uris of interest... | Slicer parses XML and presents list of URIs, researcher2 requests to download uris of interest... | ||
| − | '''send:''' curl | + | '''send:''' curl $uri |
| − | '''response: | + | '''response: file contents |
'''Step4.''' Completed neurosurgical planning analysis and upload results. | '''Step4.''' Completed neurosurgical planning analysis and upload results. | ||
Create metadatadocument (XML) for individual data file, or for scene + selected data. First, post each dataset, and assemble MRML file using returned uris. Then, post or put mrml file. | Create metadatadocument (XML) for individual data file, or for scene + selected data. First, post each dataset, and assemble MRML file using returned uris. Then, post or put mrml file. | ||
| − | '''send:''' curl - | + | '''send:''' curl -X POST -H "Content-Type: application/x-xnat-metadata+xml" -H "Content-Disposition: x-xnat-metadata; filename=\"$filename\"" -d @$metadata.xml $srv/data |
| − | '''response:''' | + | '''response:''' URI of posted file (as message body, also in Location: HTTP header) |
| − | '''send:''' curl -T | + | '''send:''' curl -T $filepath $url |
| − | '''response:''' | + | '''response:''' none |
'''Step5.''' In IGT, download scene for patient (subject) into slicer. | '''Step5.''' In IGT, download scene for patient (subject) into slicer. | ||
Query for a subject and query for available scene files... | Query for a subject and query for available scene files... | ||
| − | '''send:''' curl | + | '''send:''' curl $srv/search?subject=$subj&fileType=MRML |
'''response:''' xml containing URIs of all MRML files | '''response:''' xml containing URIs of all MRML files | ||
Slicer parses XML and presents list of MRML files, and clinican selects one with $uri. | Slicer parses XML and presents list of MRML files, and clinican selects one with $uri. | ||
| − | '''send:''' curl | + | '''send:''' curl -T $local_file $uri |
| − | '''response:''' ( | + | '''response:''' none |
| + | |||
| + | Clinician cuts network link to remote repository (all files are locally cached so OR system is self sufficient). | ||
| + | |||
| + | ''Demo install at SPL'' | ||
| − | + | * code is in /projects/birn/pieper/xnat | |
| + | * server is running on bobby on port 8000 | ||
| + | * example commands: | ||
| + | curl http://bobby:8000/search | ||
== TO-DO items for XND side== | == TO-DO items for XND side== | ||
* Send example metadata XML file (and script that writes it?) to Slicer dev. | * Send example metadata XML file (and script that writes it?) to Slicer dev. | ||
* Provide some info on webservices API as it develops | * Provide some info on webservices API as it develops | ||
| + | |||
| + | XML message schemas available in file server repository (see root URL below): | ||
| + | src/main/resources/xnat-labels.xsd | ||
| + | src/main/resources/xnat-metadata.xsd | ||
| + | src/main/resources/xnat-search.xsd | ||
| + | |||
| + | HTML description of services available here: | ||
| + | src/site/methods.html | ||
| + | |||
* Add credentialing on XND before a server is set up in a public place (consistent with XNAT E) | * Add credentialing on XND before a server is set up in a public place (consistent with XNAT E) | ||
* Establish public (read-only?) access to source code of XND web services and visual client, in version control system; | * Establish public (read-only?) access to source code of XND web services and visual client, in version control system; | ||
| + | |||
| + | Web services/file server Subversion repository: http://nrg.wustl.edu/svn/xnat-engine/server/trunk | ||
| + | |||
* command-line server style version of file server repository | * command-line server style version of file server repository | ||
| + | |||
| + | Available here: http://nrg.wustl.edu/projects/XND | ||
| + | |||
| + | Unpack the archive and read the documentation for server setup instructions. And see [[image:XNAT-FileServer-HOWTO.txt |this file for usage examples]]. | ||
| + | |||
* command-line script for using XND rules for importing data in XND database? | * command-line script for using XND rules for importing data in XND database? | ||
* turn on multiple remote repositories in the client interface, and more convenient/automated upload/download with XND client | * turn on multiple remote repositories in the client interface, and more convenient/automated upload/download with XND client | ||
| Line 92: | Line 121: | ||
=== Other Topics === | === Other Topics === | ||
| − | Misha to meet with Bernhard about deidentification algorithms for full body CT scans. | + | Misha to meet with Bernhard about deidentification algorithms for full body CT scans. ''Done - Misha to coordinate with Dan'' |
=Use-case scenarios= | =Use-case scenarios= | ||
| Line 112: | Line 141: | ||
== Use scenario 3: saving MRML files to hosted repository == | == Use scenario 3: saving MRML files to hosted repository == | ||
| − | [ | + | [https://www.slicer.org/wiki/Slicer3:XCEDE_use_cases#Slicer_use-case_.230:_Demo_we_would_like_to_show_for_BIRN described here] (similar to above). |
=== notes === | === notes === | ||
| Line 122: | Line 151: | ||
* what if a MRML scene contains data from multiple subjects, or multiple experiments, or multiple projects? How to upload this? | * what if a MRML scene contains data from multiple subjects, or multiple experiments, or multiple projects? How to upload this? | ||
| − | (Link to XNAT Desktop [ | + | (Link to XNAT Desktop [https://www.slicer.org/wiki/Slicer3:XND tutorial] for Slicer.) |
= some design & usability stuff: = | = some design & usability stuff: = | ||
Latest revision as of 18:07, 10 July 2017
Home < Events:August2008XNATDesktopPlans for the Integration of XNAT desktop and Slicer
Contents
Visit of XNAT team to Boston
- Tentative Date: August 21-22nd
- Location: 1249 Boylston Street, 2nd floor demo room Directions to 1249 Boylston
- Agenda/Goals
- Attempt to get base use case worked through
- upload files to XND
- query for them
- download them as a group
- modify them in slicer
- re-upload new files with tags
- Thursday 10am Eastern discussion with Dan Blezek about intersection with XCEDE web services implementation.
- Thursday 11pm Eastern XND Demo and discussion of local use case (C-F, Johanna, Monika)
- Attempt to get base use case worked through
- XND User Interface Review with Wendy
- People: Misha, Kevin, Wendy, Nicole, Steve, Ron (Thursday)
Plan for supporting initial use case
- Demonstrate this functionality at BIRN meeting in October
- First tests to be done by first week in September
- This plan to be presented to the NA-MIC Engineering tcon on Thursday August 28 at 3pm eastern.
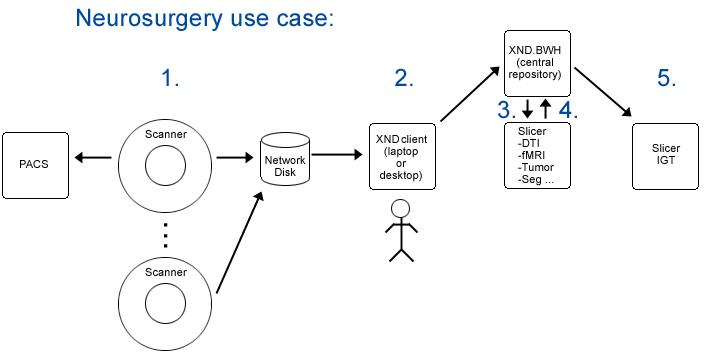
Step1. Subject is scanned and data is pushed to a networked disk (and referenced by a PACS system) automatically.
Step2. Researcher1 uses an XND client (on a laptop or a desktop system) to mark up subject data and to upload it (via drag and drop) to the central XND repository xnd.bwh.harvard.edu.
Step3. Researcher2 opens slicer and queries XND webservices on svr = http://xnd.bwh.harvard.edu:8081 for a list of subjects.
Query for all subjects on system...
send: curl $srv/search??subject response: xml for all subjects in xnd.bwh.harvard.edu
Choose a subject, query for available data...
send: curl $srv/search?subject=$subj response: xml containing URIs of all resources for the individual subject $subj
Slicer parses XML and presents list of all resources; researcher2 queries for for only DICOM data for a particular subject...
send: curl $srv/search?subject=$subj&file_type=DICOM response: xml containing URIs of all DICOM resources for subject $subj
Slicer parses XML and presents list of URIs, researcher2 requests to download uris of interest...
send: curl $uri response: file contents
Step4. Completed neurosurgical planning analysis and upload results.
Create metadatadocument (XML) for individual data file, or for scene + selected data. First, post each dataset, and assemble MRML file using returned uris. Then, post or put mrml file.
send: curl -X POST -H "Content-Type: application/x-xnat-metadata+xml" -H "Content-Disposition: x-xnat-metadata; filename=\"$filename\"" -d @$metadata.xml $srv/data response: URI of posted file (as message body, also in Location: HTTP header) send: curl -T $filepath $url response: none
Step5. In IGT, download scene for patient (subject) into slicer.
Query for a subject and query for available scene files...
send: curl $srv/search?subject=$subj&fileType=MRML response: xml containing URIs of all MRML files
Slicer parses XML and presents list of MRML files, and clinican selects one with $uri.
send: curl -T $local_file $uri response: none
Clinician cuts network link to remote repository (all files are locally cached so OR system is self sufficient).
Demo install at SPL
- code is in /projects/birn/pieper/xnat
- server is running on bobby on port 8000
- example commands:
curl http://bobby:8000/search
TO-DO items for XND side
- Send example metadata XML file (and script that writes it?) to Slicer dev.
- Provide some info on webservices API as it develops
XML message schemas available in file server repository (see root URL below): src/main/resources/xnat-labels.xsd src/main/resources/xnat-metadata.xsd src/main/resources/xnat-search.xsd
HTML description of services available here: src/site/methods.html
- Add credentialing on XND before a server is set up in a public place (consistent with XNAT E)
- Establish public (read-only?) access to source code of XND web services and visual client, in version control system;
Web services/file server Subversion repository: http://nrg.wustl.edu/svn/xnat-engine/server/trunk
- command-line server style version of file server repository
Available here: http://nrg.wustl.edu/projects/XND
Unpack the archive and read the documentation for server setup instructions. And see File:XNAT-FileServer-HOWTO.txt.
- command-line script for using XND rules for importing data in XND database?
- turn on multiple remote repositories in the client interface, and more convenient/automated upload/download with XND client
- More automated way to manage directories with changing content, and options for routing to other repositories
- Client interface usability improvements (as suggested by Wendy)
TO-DO items for Slicer side
- download data from XND to slicer
- Done, from localhost
- put data to XND from Slicer
- Working on slicer "Publish to XND" test
- post data to XND from Slicer
- Work on slicer "Post to XND" test
- better integrate remote/local load&save options in Slicer.
Questions
- vision for XNAT enterprise and XNAT desktop (longterm, how do they relate, how tightly are they coupled, and what should we use and when?)
Questions
- not sure how to regard XND (and or versus) XNATenterprise with our use cases -- where to focus development on Slicer & XNATenterprise, or Slicer & XND, etc. We would like to develop a clean implementation that meets our basic use cases, and that scales reasonably well.
Other Topics
Misha to meet with Bernhard about deidentification algorithms for full body CT scans. Done - Misha to coordinate with Dan
Use-case scenarios
Use scenario 0: (things about some external examples we like)
Use Scenario 1: fluid use of XND with Slicer
- I am using Slicer on my laptop and would like to use XNAT desktop to keep data organized on my hard disk
Capabilities needed
- how to set up when I download slicer?
- I am upgrading Slicer do I need to upgrade xnat as well?
- I worked today on a case. I would like to add my derived data and new mrml file to the existing stuff.
- I would like to upload stuff that is currently in Slicer
- I would like to download a mrml file and all associated data files.
Use scenario 3: saving MRML files to hosted repository
described here (similar to above).
notes
- StoreXAR doesn't currently have the functionality in the the use case described (getting xnat's uri back once a file is uploaded).
- Tim's suggested workaround for now: separately request a catalog of all files for a given session id. This is similar to what would be returned by StoreXAR as described in the use case.
- log into central.xnat.org and click this link to see catalog
- There's not currently a commandline executable -- we will have to write this.
- The HTTP Session ID needs to be attached to the HTTP Request.
- what if a MRML scene contains data from multiple subjects, or multiple experiments, or multiple projects? How to upload this?
(Link to XNAT Desktop tutorial for Slicer.)
some design & usability stuff:
word doc with XND and some xnat central design notes
technical questions:
- What are preferred ways to access web services – such as e.g. shell scripts calling cURL, some preferred C++ web client, Python or Java programs?
- What are requirements for XNAT File Repository web services security model?
- What would be a typical workload for various types of queries to XND (XNAT Desktop) repository - including seach, file put and file get?
- Currently, XNAT Desktop supports single file manipulation. Should it, (and if it should, then to what extent?) support treating a set of files as a single 'bundle' (for instance, "MRML file and all associated files")? What criteria should it support to automatically identify a set of files as belonging to one bundle?
- Will XNAT Desktop have to use Slicer ontology for basic tags (Project, Subject, Experiment, Scan) or will Slicer adopt, to some extent, XNAT ontology? (XNAT ontology is required to be used by Desktop when interacting with hosted XNAT Enterprise server).
Advantage Windows experience
Reference
http://slicer.spl.harvard.edu/slicerWiki/index.php/Slicer3:XCEDE_use_cases#XCEDE_REST_Interface
Comments from Misha
from Aug 18 2008
What do we have to show, from the time of last demo:
- XNAT File repository server supporting newly developed file repository REST-style Web Services, and (finally) integrated with client-server visual interface (XNAT Desktop a.k.a. XND).
- Kevin will demonstrate types of queries currently supported by Web Services, File Repository database structure and provide detail on requirements for development of Web Services clients;
- Within this scope, I can also demonstrate ontology generation approaches used by XND (which were mentioned before, DICOM rule and
regular expression rule) - most importantly, these rules are used to generate XNAT ontology from arbitrary hierarchy of files and folders.
- Go over the upload to XNAT Central, using last release of XND visual interface with improved upload capability.
Points of discussion
- We’d like to see a detailed use case for the Slicer integration, so that we could go over missing pieces in more detail. Some of the missing/unclear pieces are listed below, but it will be crucial to have a list of possible pitfalls, as complete as possible.
- Typical use of XNAT File Repository: will it be used as a local repository for Slicer projects, a hosted research data archive, a remote/back-up/secondary repository for cross-institutional data sharing, anything else?
- Prefedded way of accessing web services – such as e.g. shell scripts calling cURL, some preferred C++ web client, Python or Java programs – to test against most probable client platforms.
- Scaling: anticipated density of Web Services requests of each type (i.e. various types of search (file search, metadata search), file put / file get), from a typical Slicer client;
- Resource access control (resource-based) / session based user authentication model, other sequrity issues? Preferred security model (or at least requirements to the security model)?
- Use scenarios for XND client: standalone tool for tagging data and transferring between centralized repository and/or local users; PACS worklist-like interface for navigating DICOM data/launching viewing tools for clinical radiologists; anything else?
- Most desired features of the XND client: metadata organization/control, search/filtering features, interface organization from user perspective
- Support for Slicer ontologies, ontology conversion, ontology migration issues which will arise as soon as existing data archive will need to be converted to XNAT file repository;
- other suggestions/critiques/clarifications?