Mbirn: Slicer:DTMRI
Contents
Information about Slicer's DTMRI Module
Getting Started with the DTMRI Module
Opening DTMRI Data from the Command Line
Open an MS-DOS command window:
Go to "Start" Click "run..." In the text field type "cmd" Click "OK".
Navigate to the directory where the slicer2-win32.exe file is saved and type the following command:
>slicer2-win32.exe --no-threads --load-dicom <path/to/DTMRI/data>
A window appears indicating that Slicer is an experimental software package.
Clicking "OK" binds you to the license agreement which can be found at www.slicer.org.
Click "OK" to accept the agreement and open Slicer.
As Slicer opens, a tkcon window will appear. This is a log of Slicer's activities. Minimize this window.
Slicer Interface
For help with Slicer basics, try:
www.slicer.org; User's Guide
or Slicer Powerpoint Tutorial
When Slicer is ready to use, there will be two Slicer windows open on your screen (in addition to the tkcon window).
One is the Slicer 2.5-dev window ("Menu window" from now on), which looks like this:
The other is the Viewer window, which looks like this:
Converting DTI Data
If the data is mosaiced, the box in the lower left corner will contain several rows and columns of DTI images (see image above). Slicer has a tool that will convert mosaiced data so that individual slices can be viewed in the axial, sagittal and coronal planes.
In order to de-mosaic (convert) the data, go to the Menu. Click the "More:" button
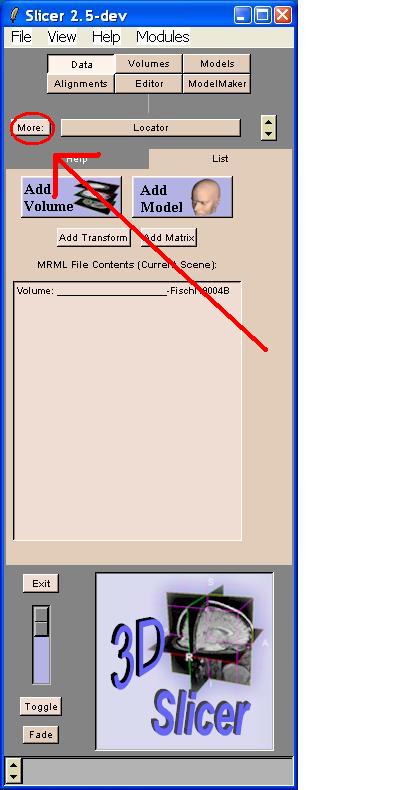
A drop down menu will appear. Choose DTMRI from this menu by clicking it.
This brings you to the DTMRI module.
About 1/4 of the way down the Menu window, there is a row of tabs.
The DTMRI module opens to the "Input" tab.
To convert your mosaiced data, click the "Convert" tab.
Within the "Convert" tab, there is a drop-down menu for "Input Volume:"
The default is the subject data that is currently open.
Do not change the default.
Under the "Input Volume" menu is the "Protocol:" menu.
The default is "None".
Change "None" to the protocol that describes the acquisition parameters of the data.
Click the "Convert Volume" button.
The "Protocol:" menu and "Convert Volume" button are indicated below.
Some error messages will appear in a vtkOutputWindow. This is normal.
The error log ends with the word "Destructing!"
Once that appears, close the vtkOutputWindow by clicking the X in the upper right corner of that window.
- WARNING: If you close the error log before "Destructing!", the program will close
and you will need to start over.
Now there are 3 diffusion tensor images on the bottom of the Viewer window.
The axial view will appear on the left, the sagittal view will appear in the center and the coronal view will appear on the right.
Once the volumes have been loaded, the views can be customized.
The default view is "Normal" view, with the 3D view at the top and the 2D planes at the bottom.
To change the View, Click "View" from the menu bar to specify the configuration of the images.
Fractional Anisotropy
Fractional Anisotropy of the Full Volume
To obtain a Fractional Anisotropy Volume from a DTI data set: Click the "More:" button

A drop down menu will appear. Choose DTMRI from this menu by clicking it.
This brings you to the DTMRI module. About 1/4 of the way down the Menu window, there is a row of tabs.
The DTMRI module opens to the "Input" tab.
If the "Scalars" tab is not visible, click on the "More..." tab.
Click on the "Scalars" tab. Set Scale Factor to 1000.
Click "Apply". As you move the mouse over any of the 2D views you will see the underlying pixel values in the lower left corner of each image. By default, the Fractional Anisotropy value is selected as the "background" volume, so the the value that you see as "Bg" is the Fractional Anisotropy multiplied by the specified scale factor (1000 in this case).
Fractional Anistropy of an ROI
To find the FA in a particular region of interest, create a labelmap of the ROI.
Go to the Editor Module
The Editor Module automatically opens to the "Volumes" tab.
Leave "Original Grayscale:" as the default and "Working Labelmap:" as "NEW"
Choose a "Descriptive Name" that describes the labelmap that will be created.
*Note: The descriptive name cannot contain spaces.
Click "Start Editing".
The "Effects" tab opens.
Click the "Draw" button.
The "Details" tab opens.
The default color for a new labelmap is "Skin".
If you want to change the label color, click "Output:"
A window appears from which you can select a color. Click on the desired color from the color palette. (Note: Do not choose Black or 0.)
Click on a number in the "Labels" menu (any number listed will work).
Change other options in the Details tab where appropriate.
"Render" determines whether edits apply to one slice, 3 slices, or the entire 3D volume.
"Shape" determines how edits will be interpreted.
- If the shape is "Polygon", the points that are drawn will be
connected and the interior of the polygon created will be
filled
when the user clicks "Apply". - If the shape is "Lines", the points will be connected in a linear fashion.
- If the shape is "Points", the edits will not be connected or filled.
Use the left button on the mouse to specify the ROI.
Use "Delete Selected" to undo edits one point at a time.
Use "Delete All" do delete all edits since the edits were last applied.
Click "Apply" to set the ROI.
Add new areas to the labelmap by specifying them and clicking "Apply".
To delete part of an ROI after the edits have been applied, change the "Output" label number to 0.
Use Polygon to outline the unwanted area.
Click "Apply".
This is an example of a labelmap of the corpus callosum created on an FA volume.
The label is #15 mambazo and is most easily seen in the bottom center view in the Viewer window.
Once the ROI has been sufficiently identified in the labelmap, go to the "Module" menu.
Go to "Meaurement" and then to "VolumeMath".
Click on the "Cast" button.
In the "Volume to Cast:" menu, choose "FractionalAnisotropy____..."
Leave "(not used)" as "None".
In the "Cast Output" menu, choose "Create New".
In the "Output Type" menu, choose "Short".
Click the "Run" button.
Slicer will create "VolumeMathCastResult-#".
*Note: # changes each time you run Cast and MaskStat.
Click the "MaskStat" button.
In the "Volume to Mask:" menu, choose "VolumeMathCastResult-#".
In the "Label Map:" menu, choose the Descriptive Name of the Label Map created.
In the "Masked Output:" menu, choose "Create New".
Check that the "Mask:" color identifiers match those of the label map created.
In the "Output File" field, use "Browse" to specify the destination directory
and name of the text file that will be created.
This text file will contain the FA data for the ROI.
Click "Run".
A pop-up window will appear on the screen containing:
- The minimum FA within the ROI (multiplied by a factor of 1000).
- The maximum FA within the ROI (multiplied by a factor of 1000).
- The mean FA within the ROI (multiplied by a factor of 1000).
- The standard deviation of the FA within the ROI.
Click "OK". Slicer creates a text file in the specified directory with the same data.
Tractography
Synthesizing Axon Tracts
Slicer synthesizes tracts from a chosen seed point.
The algorithm works as follows. From the seed point, Slicer determines the direction of maximum diffusion
(ie the direction in which the Fractional Anisotropy is closest to 1.0).
A fiber (or tract) is represented graphically by a cylindrical tube in the 3D view of the viewer window.
Slicer continues synthesizing a given tract until one of the following stop criterion is reached:
- The tract reaches a maximum length (measured in mm).
- The tract makes a sudden change in direction.
- The Fractional Anisotropy drops below a certain threshold.
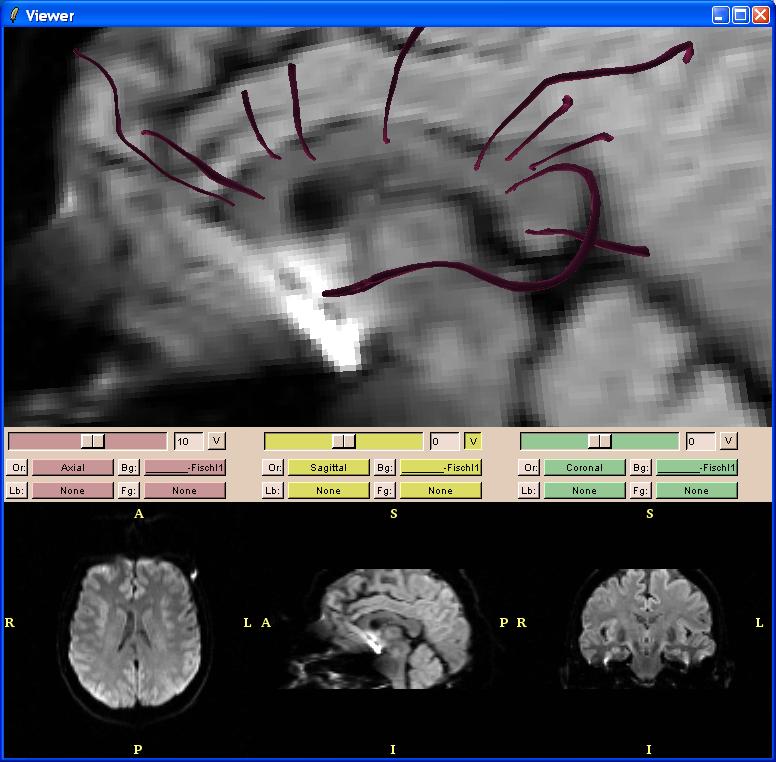
Here is an example of 10 tracts synthesized from 10 manually-defined seed points on the sagittal view of the corpus callosum:
The stop criteria can be modified by going to the Display tab ("Disp" or "Display").
Within the Display Tab, click on the Tractography tab.
In "Visualization Menu:" choose "Tracts".
To modify the maximum propagation length of the axon, change the value in the "Max Length:" field.
Tractography will stop synthesizing the axon when it reaches this length.
To modify the threshold for change in tract direction,
change the value in the "Curvature Threshold:" field (the 2nd field from the bottom).
To modify the minimum Fractional Anisotropy, change the value in the "FA Threshold:" field.
Synthesizing Axon Tracts Individually
Axon tracts can be synthesized when in the DTMRI module.
In the Viewer window, click inside the view where seed points will be specified to activate that window.
Tracts can be synthesized in the axial, sagittal, coronal and 3D views.
Hover the mouse over the area where the axon is to be synthesized.
Press the "s" key.
Repeat as desired.
To delete synthesized tracts, hover the mouse over the tract to be deleted and press the "d" key.
To change the color of your tracts:
In the DTMRI module, click on the Display tab ("Disp" or "Display")
Go to the "Tractography" tab.
The default color for new tracts is "Skin".
To change the label color, click "Color:"
A window appears from which you can select a color.
Click on the desired color from the color palette. (Note: Do not choose Black or 0.)
Once the desired color is selected (it will appear to the right of "Color:"),
use "s" and "d" as explained above.
Synthesizing Tracts from a Region Of Interest
Slicer can synthesize tracts from a user-defined ROI.
To create an ROI from DTI volumes:
Go to the Editor Module
The Editor Module automatically opens to the "Volumes" tab.
Leave "Original Grayscale:" as the default and "Working Labelmap:" as "NEW"
Choose a "Descriptive Name" that describes the labelmap that will be created.
*Note: The descriptive name cannot contain spaces.
Click "Start Editing".
The "Effects" tab opens.
Click the "Draw" button.
The "Details" tab opens.
The default color for a new labelmap (and the tracts that will be synthesized from it) is "Skin".
If you want to change the label color, click "Output:"
A window appears from which you can select a color. Click on the desired color from the color palette. (Note: Do not choose Black or 0.)
Click on a number in the "Labels" menu (any number listed will work).
Change other options in the Details tab where appropriate.
"Render" determines whether edits apply to one slice, 3 slices, or the entire 3D volume.
"Shape" determines how edits will be interpreted.
- If the shape is "Polygon", the points that are drawn will be
connected and the interior of the polygon created will be
filled
when the user clicks "Apply". - If the shape is "Lines", the points will be connected in a linear fashion.
- If the shape is "Points", the edits will not be connected or filled.
Use the left button on the mouse to specify the ROI.
Use "Delete Selected" to undo edits one point at a time.
Use "Delete All" do delete all edits since the edits were last applied.
Click "Apply" to set the ROI.
Add new areas to the labelmap by specifying them and clicking "Apply".
To delete part of an ROI after the edits have been applied, change the "Output" label number to 0.
Use Polygon to outline the unwanted area.
Click "Apply".
This is an example of a labelmap of the corpus callosum created on a DTI volume.
The label is #2 skin and is most easily seen in the bottom center view in the Viewer window.
Once the ROI has been sufficiently identified in the labelmap, go back to the DTMRI module.
Do this by clicking on the "DTMRI" button (above the tabs in the Menu window)
or by choosing "DTMRI" from the "More:" menu.
Go to the "Display" tab.
Inside the panel, go to the "Tractography" tab.
In "Visualization Menu:" choose "AutoTracts".
In the "ROI Labelmap:" choose the Descriptive Name specified when the labelmap was created.
Click "Seed 'Tracts' in ROI".
An alert window will appear. It says,
"About to seed streamlines in all labelled voxels of volume <descriptive_name>.
This may take a while, so make sure the Tracts settings are what you want first. Go ahead?"
- Note: If you want to change the Tracts settings, go to "Visualization Menu" and choose "Tracts".
Modify as necessary.
Change "Visualization Menu" back to "Auto Tracts", specify the ROI labelmap and continue.
Click "Yes".
Slicer will grow tracts with the ROI as seed points based on the algorithm described above.
This could take a few minutes depending on the ROI and the settings.
This is an example of the tracts grown using a DTI volume to identify the corpus callosum as a labelmap:
From here, it is possible to synthesize additional axons or delete unwanted ones.
To synthesize axons:
- Click in the view where axons will be synthesized.
- Hover the mouse over the area where the axon is to be synthesized.
- Press the "s" key.
- Repeat as desired.
To delete synthesized tracts, hover the mouse over the tract to be deleted and press the "d" key.
Synthesizing Tracts from ROI's Created in Other Volumes
It is possible to create Labelmaps of ROIs from other volumes (eg. T1, FA)
and use them grow tracts from those ROIs in the DTMRI module.
Synthesizing Tracts from an Original Volume
To load another data set, go to the Data module by clicking the "Data" button.
Choose the "List" tab.
Click "Add Volume".
For information on loading volumes, go to www.slicer.org --> User's Guide --> Loading and Viewing data.
If the new volumes do not appear immediately, go to the 2D views in the Viewer window.
- At the top of each view, there are 4 menus: "Or:", "Lb:", "Bg:" and "Fg:"
- Choose the new volume from the "Bg:" menu in each of the 2D views.
To create an ROI from this volume:
Go to the Editor Module
The Editor Module automatically opens to the "Volumes" tab.
Leave "Original Grayscale:" as the default.
The "Working Labelmap:" should be "NEW".
Choose a "Descriptive Name" that describes the labelmap that will be created.
*The descriptive name cannot contain spaces.
Click "Start Editing".
The "Effects" tab opens.
Click the "Draw" button.
The "Details" tab opens.
The default color for a new labelmap (and the tracts that will be synthesized from it) is "Skin".
If you want to change the label color, click "Output:"
A window appears from which you can select a color.
Click on the desired color from the color palette. (Note: Do not choose Black or 0.)
Click on a number in the "Labels" menu (any number listed will work).
Change other options in the Details tab where appropriate.
"Render" determines whether edits apply to one slice, 3 slices, or the entire 3D volume.
"Shape" determines how edits will be interpreted.
- If the shape is "Polygon", the points that are drawn will be
connected and the interior of the polygon created will be
filled when the user clicks "Apply". - If the shape is "Lines", the points will be connected in a linear fashion.
- If the shape is "Points", the edits will not be connected or filled.
Use the left button on the mouse to specify the ROI.
Use "Delete Selected" to undo edits one point at a time.
Use "Delete All" do delete all edits since the edits were last applied.
Click "Apply" to set the ROI.
Add new areas to the labelmap by specifying them and clicking "Apply".
To delete part of an ROI after the edits have been applied, change the "Output" label number to 0.
Use Polygon to outline the unwanted area.
Click "Apply".
This is an example of a labelmap of the corpus callosum created on an MPRage volume.
The label is #7 and is most easily seen in the bottom center view in the Viewer window.
Once the ROI has been sufficiently identified in the labelmap, go back to the DTMRI module.
Do this by clicking on the "DTMRI" button (above the tabs in the Menu window)
or by choosing "DTMRI" from the "More:" menu.
Choose the "Display" (or "Disp") tab.
Within the Display panel, go to the "Tractography" tab.
In "Visualization Menu:" choose "AutoTracts".
In "ROI Labelmap:" choose the labelmap created in the new volume.
Make sure the Label is set to the one used in created the labelmap.
Click "Seed 'Tracts' in ROI".
An alert window will appear. It says,
"About to seed streamlines in all labelled voxels of volume <descriptive_name>. This may
take a while, so make sure the Tracts settings are what you want first. Go ahead?"
- Note: If you want to change the Tracts settings, go to "Visualization Menu" and choose "Tracts".
Modify as necessary.
Change "Visualization Menu" back to "Auto Tracts", specify the ROI labelmap and continue.
Click "Yes".
Slicer will grow tracts with the ROI as seed points based on the algorithm described above.
This could take a few minutes depending on the ROI and the settings.
This is an example of the tracts grown using a T1 volume to identify the corpus callosum as a labelmap:
From here, it is possible to synthesize additional axons or delete unwanted ones.
To synthesize axons:
- Click in the view where axons will be synthesized.
- Hover the mouse over the area where the axon is to be synthesized.
- Press the "s" key.
- Repeat as desired.
To delete synthesized tracts, hover the mouse over the tract to be deleted and press the "d" key.
Synthesizing Tracts from a Scalar Volume
The process for creating one type of scalar volume, a Fractional
Anisotropy volume, is described above in Fractional Anisotropy of the Full Volume.
Other types of scalar volumes (eg Linear Measure, Relative Anisotropy) can
be created by choosing a different option from the "Create Volume:" menu
in the "Scalars" tab of the DTMRI module.
Once a scalar volume has been created, it can be used to guide synthesis of individual tracts or to create a labelmap of a region of interest.
Synthesizing Individual Tracts from a Scalar Volume
To synthesize individual tracts from a scalar volume, go to the "Display" tab in the DTMRI module.
In "Display 'Tracts':", click "On".
In "Visualization Menu:", choose "Tracts".
In the viewer window,
- Click in the view where axons will be synthesized.
- Hover the mouse over the area where the axon is to be synthesized.
- Press the "s" key.
- Repeat as desired.
To delete synthesized tracts, hover the mouse over the tract to be deleted and press the "d" key.
Synthesizing Tracts from an ROI defined in a Scalar Volume
To synthesize tracts using an ROI created from a scalar volume:
Go to the Editor Module
The Editor Module automatically opens to the "Volumes" tab.
Leave "Original Grayscale:" as the default (your scalar volume).
The "Working Labelmap:" should be "NEW".
Choose a "Descriptive Name" that describes the labelmap that will be created.
*The descriptive name cannot contain spaces.
Click "Start Editing".
The "Effects" tab opens.
Click the "Draw" button.
The "Details" tab opens.
The default color for a new labelmap (and the tracts that will be synthesized from it) is "Skin".
If you want to change the label color, click "Output:"
A window appears from which you can select a color.
Click on the desired color from the color palette. (Note: Do not choose Black or 0.)
Click on a number in the "Labels" menu (any number listed will work).
Change other options in the Details tab where appropriate.
"Render" determines whether edits apply to one slice, 3 slices, or the entire 3D volume.
"Shape" determines how edits will be interpreted.
- If the shape is "Polygon", the points that are drawn will be
connected and the interior of the polygon created will be
filled when the user clicks "Apply". - If the shape is "Lines", the points will be connected in a linear fashion.
- If the shape is "Points", the edits will not be connected or filled.
Use the left button on the mouse to specify the ROI.
Use "Delete Selected" to undo edits one point at a time.
Use "Delete All" do delete all edits since the edits were last applied.
Click "Apply" to set the ROI.
Add new areas to the labelmap by specifying them and clicking "Apply".
To delete part of an ROI after the edits have been applied, change the "Output" label number to 0.
Use Polygon to outline the unwanted area.
Click "Apply".
Once the ROI has been sufficiently identified in the labelmap, go back to the DTMRI module.
Do this by clicking on the "DTMRI" button (above the tabs in the Menu window)
or by choosing "DTMRI" from the "More:" menu.
Choose the "Display" (or "Disp") tab.
Within the Display panel, go to the "Tractography" tab.
In "Visualization Menu:" choose "AutoTracts".
In "ROI Labelmap:" choose the labelmap created in the new volume.
Make sure the Label is set to the one used in created the labelmap.
Click "Seed 'Tracts' in ROI".
An alert window will appear. It says,
"About to seed streamlines in all labelled voxels of volume <descriptive_name>. This may
take a while, so make sure the Tracts settings are what you want first. Go ahead?"
- Note: If you want to change the Tracts settings, go to "Visualization Menu" and choose "Tracts".
Modify as necessary.
Change "Visualization Menu" back to "Auto Tracts", specify the ROI labelmap and continue.
Click "Yes".
Slicer will grow tracts with the ROI as seed points based on the algorithm described above.
This could take a few minutes depending on the ROI and the settings.
Once the tracts have been synthesized, it is possible to synthesize
additional axons or delete unwanted ones.
To synthesize axons:
- Click in the view where axons will be synthesized.
- Hover the mouse over the area where the axon is to be synthesized.
- Press the "s" key.
- Repeat as desired.
To delete synthesized tracts, hover the mouse over the tract to be deleted and press the "d" key.
Using a Scalar Volume to Color Synthesized Tracts
As an alternative to manually specifying the colors of tracts created from
a scalar volume, Slicer is capable of adjusting the color of tracts based
on the scalar volume.
To use this option, go to the "Display" tab in the DTMRI module.
In "Display 'Tracts':", choose "On".
In "Visualization Menu", choose "Tracts"
"Color:" can be left as the default.
In "Color by:", choose "MultiColor".
In "Color by Volume:", choose your scalar volume from the menu.
To synthesize axons:
- Click in the view where axons will be synthesized.
- Hover the mouse over the area where the axon is to be synthesized.
- Press the "s" key.
- Repeat as desired.
To delete synthesized tracts, hover the mouse over the tract to be deleted and press the "d" key.
You'll notice that the tracts are a different color than the one specified in "Color:".
When you use this process, Slicer interpolates the scalar volume to produce the colors.
It is also possible to use the MultiColor option with tracts synthesized
from an ROI.
Once the tracts have been created from the labelmap, as described above,
go to the "Tractography" tab in the "Display" tab of the DTMRI module.
In "Visualization Menu:", choose "Tracts" In "Color by:", choose "MultiColor" In "Color by Volume:", choose the "Descriptive Name" that you specified
for your labelmap.
Slicer will update the colors of your tracts in the 3D viewer window.
Saving Tracts
Saving tracts created in the DTMRI module involves several steps.
First, save a model that contains the tracts
then save a scene containing the model with the tracts.
Saving a Model with Tracts
Once the tracts have been created, go to the Tractography Panel
in the Display Tab of the DTMRI module.
In the "Visualization Menu:" choose "SaveTracts"
The SaveTracts Panel opens.
Click the "Save tracts in model file(s)" button.
Browse to a directory, name the file and click "Save".
A message appears which indicates that Slicer has finished
writing the necessary files.
The file name is specified (eg. <myTractsModel>.xml).
Click OK.
Saving the Scene
Go to the Data module.
Click the "Add Model" button. 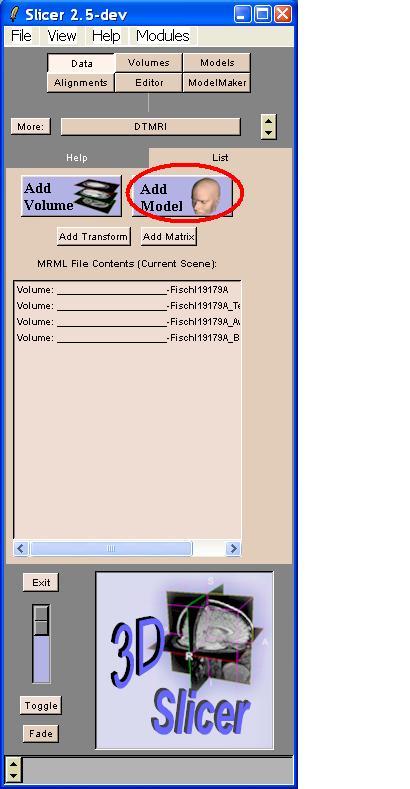
The Props Tab of the Models Module opens, which looks like this:
Click the "Browse" button.
Navigate to the directory where the model containing the tracts was saved.
Click on the file named <myTractsModel>_0.vtk
Click "Open".
On the Models Props tab, click "Apply".
The Menu screen returns to the Data module.
The "MRML File Contents (Current Scene):" window should list:
- The original DTI data
- The DTI tensor
- The DTI AvGradient
- The DTI Baseline
- The model containing tract
- Other volumes that have been added
(see Creating_ROI's_from_Other_Volumes)
Saving the scene will create a file that records the paths to
- The original DTI data
- Models
- The original data of other volumes added
The DTI tensor will need to be converted each time the data is opened.
See
The "SaveAs File" window appears.
Click "Browse". Navigate to the directory where the scene is to be saved.
Name the scene.
Click "Save".
A warning window appears to indicate that the Tensor,
AvGradient and Baseline files will not be saved as
part of the scene.
Click "Yes".
The scene is now saved as <myScene>.xml
When Slicer closes, a warning message will appear indicating that
the Tensor, AvGradient and Baseline files have not been saved.
Click "Yes" to exit anyway.
Opening Saved Tracts
With Slicer open, go to File in the Menu window.
Choose "Open Scene..."
The "Open File" screen appears.
If <myScene> appears in the window
- Click "Apply"
- The scene will open.
If <myScene> does not appear in the "Open File" screen
- Click "Browse".
- Navigate to the directory containing <myScene>.xml
- Open <myScene>.xml
- The scene will open.
Convert the DTI data (see Converting_DTI_Data.
The converted DTI data, tracts and other volumes in the scene will
be visible in the viewer windows.