Difference between revisions of "NA"
From NAMIC Wiki
m (Update from Wiki) |
m (Update from Wiki) |
||
| Line 1: | Line 1: | ||
| − | '''Objective:''' | + | '''Objective:''' We want to add various statistical measures into our PDE flows for medical imaging. This will allow the incorporation of global image information into the locally defined PDE frameowrk. |
| − | '''Progress:''' | + | '''Progress:''' We developped flows which can separate the distributions inside and outside the evolving contour, and we have also been including shape information in the flows. |
| − | + | '''Completed:''' | |
| − | + | * A statistically based flow for image segmentation, using Fast Marching | |
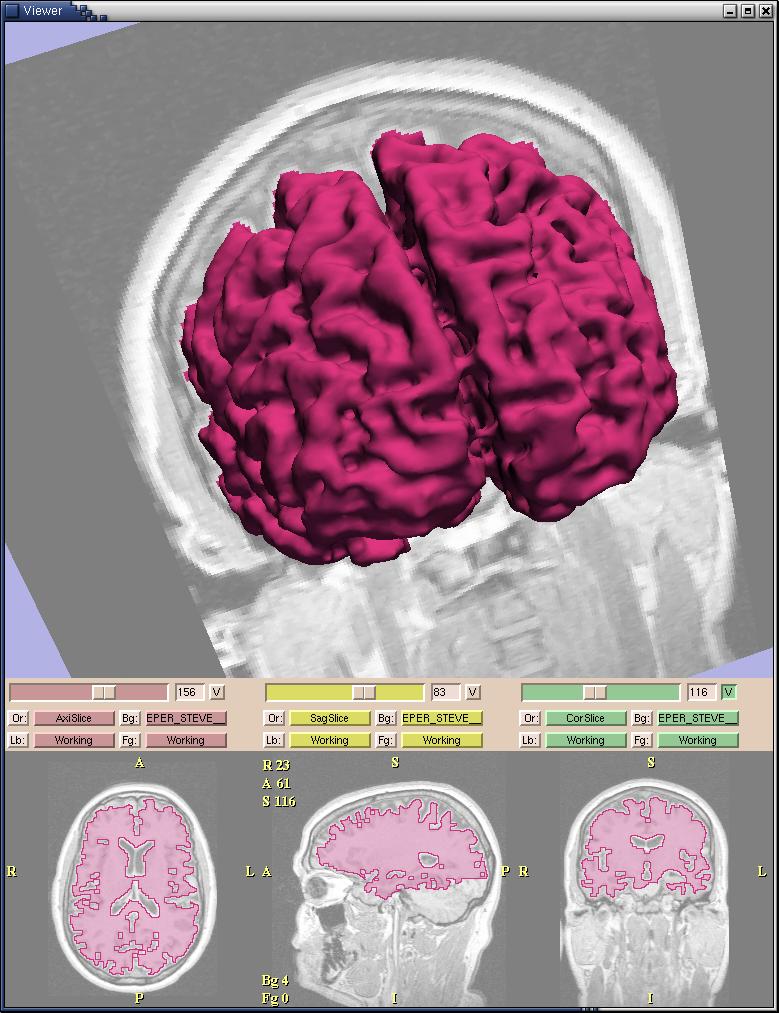
| − | + | <div class="thumb tright"><div style="width: 182px">[[Image:Gatech_SlicerModel2.jpg|[[Image:180px-Gatech_SlicerModel2.jpg|Figure 1:Screenshot from the Slicer Fast Marching module]]]]<div class="thumbcaption"><div class="magnify" style="float: right">[[Image:Gatech_SlicerModel2.jpg|[[Image:magnify-clip.png|Enlarge]]]]</div>Figure 1:Screenshot from the Slicer Fast Marching module</div></div></div> | |
| − | + | ''Code'' | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | * The code has been integrated into the Slicer | |
| + | * A user-oriented tutorial for the Fast Marching algorithm is available at:[http://www.bme.gatech.edu/groups/minerva/publications/papers/pichon.slicer.fastMarching/index.html Slicer Module Tutorial] | ||
| − | * | + | ''References:'' |
| − | + | ||
| + | * Eric Pichon, Allen Tannenbaum, and Ron Kikinis. A statistically based surface evolution method for medical image segmentation: presentation and validation. In International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), volume 2, pages 711-720, 2003. Note: Best student presentation in image segmentation award[http://www.bme.gatech.edu/groups/minerva/publications/papers/pichon-media2004-segmentation.pdf [1]] | ||
| + | |||
| + | '''Key Investigators:''' Delphine Nain, Eric Pichon, Oleg Michailovich, Yogesh Rathi, James Malcolm, Allen Tannenbaum | ||
'''Links:''' | '''Links:''' | ||
| − | + | * [[Algorithm:GATech#The_Fast_Marching_algorithm_has_been_integrated_into_the_Slicer.:GATech|GATech Tech Summary Page]] | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Revision as of 13:28, 18 December 2006
Home < NAObjective: We want to add various statistical measures into our PDE flows for medical imaging. This will allow the incorporation of global image information into the locally defined PDE frameowrk.
Progress: We developped flows which can separate the distributions inside and outside the evolving contour, and we have also been including shape information in the flows.
Completed:
- A statistically based flow for image segmentation, using Fast Marching
Code
- The code has been integrated into the Slicer
- A user-oriented tutorial for the Fast Marching algorithm is available at:Slicer Module Tutorial
References:
- Eric Pichon, Allen Tannenbaum, and Ron Kikinis. A statistically based surface evolution method for medical image segmentation: presentation and validation. In International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), volume 2, pages 711-720, 2003. Note: Best student presentation in image segmentation award[1]
Key Investigators: Delphine Nain, Eric Pichon, Oleg Michailovich, Yogesh Rathi, James Malcolm, Allen Tannenbaum
Links: