Difference between revisions of "NA-MIC/Projects/Collaboration/MGH RadOnc"
| (38 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| + | __NOTOC__ | ||
| + | |||
| + | ===Adaptive RT for Head and Neck=== | ||
| + | |||
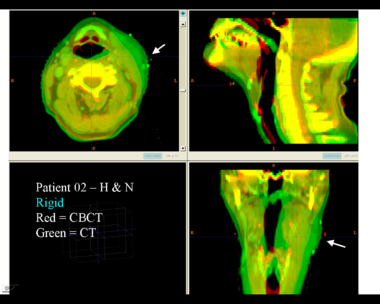
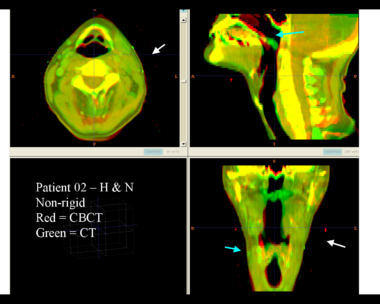
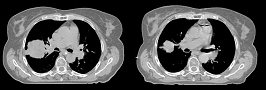
| + | This first example shows the kind of anatomic changes that can occur in head and neck cancer. The pre-treatment scan is in green, and the mid-treatment scan is in red. The image on the left is the rigid registration, on the right is deformable registration. | ||
| + | |||
{| | {| | ||
| − | |[[Image: | + | |[[Image:RadOnc_HN2_rigid.png|thumb|380px|Rigid registration]] |
| − | |[[Image: | + | |[[Image:RadOnc_HN2_deformable.png|thumb|380px|Deformable registration]] |
|} | |} | ||
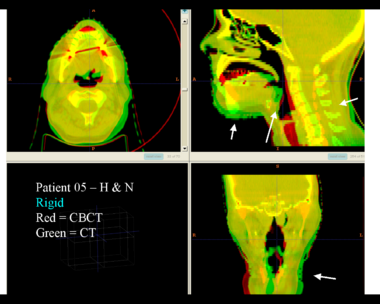
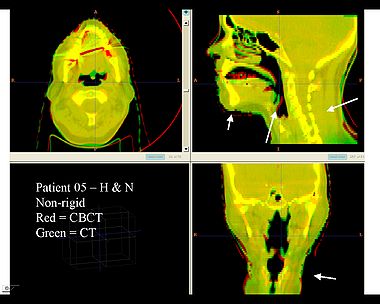
| − | + | Here is another pertinent example for head and neck. In axial view, there appears to be some weight loss. Note the change in positioning of the mandible, and also the twisting of the cervical spine between scans. Also note the strong CT artifacts caused by dental fillings. In both examples, registration of the soft palette is worse using deformable registration than rigid registration, probably due to these artifacts. | |
| + | |||
| + | {| | ||
| + | |[[Image:RadOnc_HN5_rigid.png|thumb|380px|Rigid registration]] | ||
| + | |[[Image:RadOnc_HN5_deformable.jpeg|thumb|380px|Deformable registration]] | ||
| + | |} | ||
| + | |||
| + | |||
| + | |||
| + | ===Adaptive RT for Thorax=== | ||
| + | |||
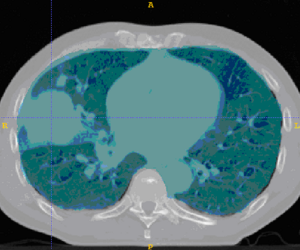
| + | This example shows anatomic change in the thorax. The patient has a collapsed left lower lobe in the pre-treatment scan (top), which has recovered in the mid-treatment scan (bottom). Notice there is some kind of fluid accumulation below the collapsed lung. | ||
| + | |||
| + | {| | ||
| + | |[[Image:RadOnc_LU1.png|thumb|380px|Lung cancer 1]] | ||
| + | |} | ||
| − | + | Here is another example, showing the magnitude of tumor regression that can occur during treatment. (Sorry, low resolution image). | |
| − | |||
| − | |||
| + | {| | ||
| + | |[[Image:RadOnc_LU2.png|thumb|380px|Lung cancer 2]] | ||
| + | |} | ||
| − | + | Thorax is a special case. Patient images are acquired using 4D-CT, and radiation treatment plan can be evaluated at each breathing phase. The volumes are aligned using deformable registration, and radiation dose from each phase is accumulated into a reference phase (e.g. exhale). This process is called "4D treatment planning." | |
| − | + | Deformable registration of the phases within a 4D-CT is considered "easy". The reason for this is: | |
| − | + | # Single-session imaging, so patient is already co-registered | |
| − | + | # Single-session imaging, so no anatomic change | |
| + | # High contrast of vessels against lung parenchema | ||
| − | + | However, the sliding of the lungs against the chest wall is difficult to model. We sometimes segment the images at the pleural boundary. This allows us to separate the moving set of organs from the non-moving set, which are registered separately. Ideally we would always do this, but segmentation is manual and therefore we usually skip this step. | |
| − | + | {| | |
| + | |[[Image:RadOnc_LU3.png|thumb|300px|Segmentation at pleural boundary]] | ||
| + | |[[Image:RadOnc_LU4_before.png|thumb|300px|4D-CT phases]] | ||
| + | |[[Image:RadOnc_LU4_after.png|thumb|300px|Registration of 4D-CT phases]] | ||
| + | |} | ||
| − | + | ===General Discussion of Registration=== | |
| − | |||
| − | + | Deformable registration is still not as reliable as it should be. Here are some of my complaints: | |
| − | + | # Image acquisition has residual artifacts which cause unrealistic deformations (dental artifacts in H&N, motion artifacts in thorax). | |
| + | # Registration algorithms are not always robust, and require experimentation and tuning. | ||
| + | # Validation of registration results is not easy, since there are inadequate tools. | ||
| + | # In thorax, temporal regularization is generally not done, because of slow algorithms and large memory footprints. | ||
| + | # For cone-beam CT (and sometimes MR), intensity values are not globally stable. The most common suggestion is adaptive histogram equalization, but isn't there a better way? | ||
| + | # Interactive tools to repair (or reinitialize) registration are virtually non-existent. | ||
| − | + | ===Segmentation=== | |
| − | + | Rad Onc departments use interactive segmentation every day for both research and patient care. | |
| − | + | Prior to treatment planning, the target and critical structures are delineated in CT. The current state of the art is manual segmentation in axial view. A outline tool, used delineate the boundary, is generally prefered over a paintbrush tool that fills pixels. Commercial products generally support some subset of the following tools to assist the operator. | |
| − | + | # contour interpolation between slices | |
| + | # boundary editing | ||
| + | # mixed axial/coronal/sagittal drawing | ||
| + | # livewire or intelligent scissors | ||
| + | # drawing constraints (e.g. constraints on volume overlap/distance) | ||
| + | # post-processing tools to nudge or smooth the boundary | ||
| − | + | There are many opportunities for using computer vision algorithms to improve interactive segmentation. For example, using prior models of shape and intensity to improve interpolation. Automatic segmentation also exists is several commercial products, each with impressive demos. We have GE Adv Sim software which does model-based segmentation of structures such as spinal cord, lens & optic nerve, liver. For H&N, atlas-based segmentation is the most popular approach. | |
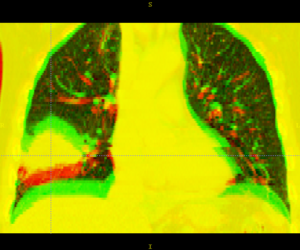
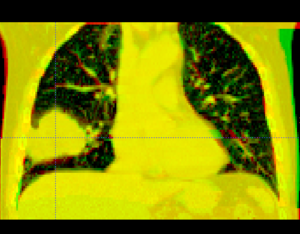
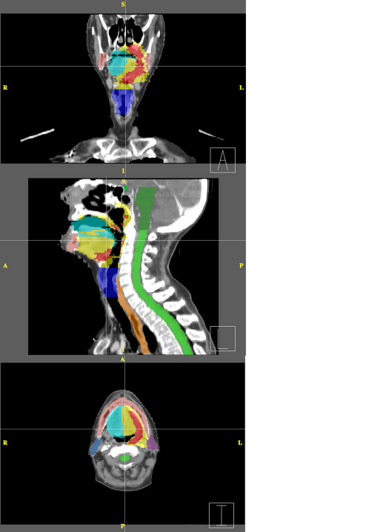
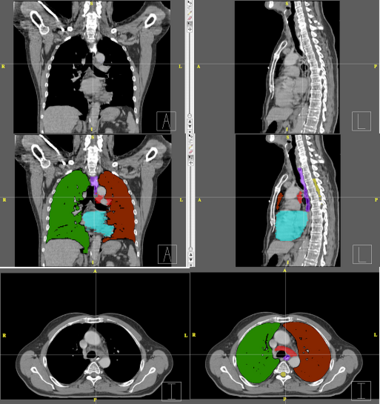
| − | + | Below are examples of segmentations of targets and critical structures for head and neck, and thorax. These structures (or similar) would be manually delineated for every patient who gets 3D planning. | |
| + | {| | ||
| + | |[[Image:RadOnc_HN_seg.png|thumb|380px|Head and Neck segmentation]] | ||
| + | | | ||
| + | {| class="wikitable" | ||
| + | |- | ||
| + | ! optic chiasm | ||
| + | ! med green | ||
| + | |- | ||
| + | ! brain stem | ||
| + | ! dim green | ||
| + | |- | ||
| + | ! spinal cord | ||
| + | ! bright green | ||
| + | |- | ||
| + | ! left parotid | ||
| + | ! violet | ||
| + | |- | ||
| + | ! right parotid | ||
| + | ! dim blue | ||
| + | |- | ||
| + | !oral cavity | ||
| + | ! cyan | ||
| + | |- | ||
| + | ! mandible | ||
| + | ! pink | ||
| + | |- | ||
| + | ! larynx | ||
| + | ! bright blue | ||
| + | |- | ||
| + | ! esophagus | ||
| + | ! orange | ||
| + | |- | ||
| + | ! (target) | ||
| + | ! red | ||
| + | |- | ||
| + | ! (target) | ||
| + | ! yellow | ||
| + | |} | ||
| + | |} | ||
| − | = | + | {| |
| − | + | |[[Image:RadOnc_LU_seg.png|thumb|380px|Thorax segmentation]] | |
| − | + | | | |
| − | + | {| class="wikitable" | |
| − | + | |- | |
| + | ! left lung | ||
| + | ! dark red | ||
| + | |- | ||
| + | ! right lung | ||
| + | ! green | ||
| + | |- | ||
| + | ! esophagus | ||
| + | ! violet | ||
| + | |- | ||
| + | ! heart | ||
| + | ! cyan | ||
| + | |- | ||
| + | ! cord | ||
| + | ! yellow | ||
| + | |- | ||
| + | ! (target) | ||
| + | ! light red | ||
| + | |} | ||
| + | |} | ||
Latest revision as of 22:26, 8 June 2009
Home < NA-MIC < Projects < Collaboration < MGH RadOnc
Adaptive RT for Head and Neck
This first example shows the kind of anatomic changes that can occur in head and neck cancer. The pre-treatment scan is in green, and the mid-treatment scan is in red. The image on the left is the rigid registration, on the right is deformable registration.
Here is another pertinent example for head and neck. In axial view, there appears to be some weight loss. Note the change in positioning of the mandible, and also the twisting of the cervical spine between scans. Also note the strong CT artifacts caused by dental fillings. In both examples, registration of the soft palette is worse using deformable registration than rigid registration, probably due to these artifacts.
Adaptive RT for Thorax
This example shows anatomic change in the thorax. The patient has a collapsed left lower lobe in the pre-treatment scan (top), which has recovered in the mid-treatment scan (bottom). Notice there is some kind of fluid accumulation below the collapsed lung.
Here is another example, showing the magnitude of tumor regression that can occur during treatment. (Sorry, low resolution image).
Thorax is a special case. Patient images are acquired using 4D-CT, and radiation treatment plan can be evaluated at each breathing phase. The volumes are aligned using deformable registration, and radiation dose from each phase is accumulated into a reference phase (e.g. exhale). This process is called "4D treatment planning."
Deformable registration of the phases within a 4D-CT is considered "easy". The reason for this is:
- Single-session imaging, so patient is already co-registered
- Single-session imaging, so no anatomic change
- High contrast of vessels against lung parenchema
However, the sliding of the lungs against the chest wall is difficult to model. We sometimes segment the images at the pleural boundary. This allows us to separate the moving set of organs from the non-moving set, which are registered separately. Ideally we would always do this, but segmentation is manual and therefore we usually skip this step.
General Discussion of Registration
Deformable registration is still not as reliable as it should be. Here are some of my complaints:
- Image acquisition has residual artifacts which cause unrealistic deformations (dental artifacts in H&N, motion artifacts in thorax).
- Registration algorithms are not always robust, and require experimentation and tuning.
- Validation of registration results is not easy, since there are inadequate tools.
- In thorax, temporal regularization is generally not done, because of slow algorithms and large memory footprints.
- For cone-beam CT (and sometimes MR), intensity values are not globally stable. The most common suggestion is adaptive histogram equalization, but isn't there a better way?
- Interactive tools to repair (or reinitialize) registration are virtually non-existent.
Segmentation
Rad Onc departments use interactive segmentation every day for both research and patient care. Prior to treatment planning, the target and critical structures are delineated in CT. The current state of the art is manual segmentation in axial view. A outline tool, used delineate the boundary, is generally prefered over a paintbrush tool that fills pixels. Commercial products generally support some subset of the following tools to assist the operator.
- contour interpolation between slices
- boundary editing
- mixed axial/coronal/sagittal drawing
- livewire or intelligent scissors
- drawing constraints (e.g. constraints on volume overlap/distance)
- post-processing tools to nudge or smooth the boundary
There are many opportunities for using computer vision algorithms to improve interactive segmentation. For example, using prior models of shape and intensity to improve interpolation. Automatic segmentation also exists is several commercial products, each with impressive demos. We have GE Adv Sim software which does model-based segmentation of structures such as spinal cord, lens & optic nerve, liver. For H&N, atlas-based segmentation is the most popular approach.
Below are examples of segmentations of targets and critical structures for head and neck, and thorax. These structures (or similar) would be manually delineated for every patient who gets 3D planning.
|
|