Difference between revisions of "NA-MIC/Projects/Collaboration/MGH RadOnc"
| Line 1: | Line 1: | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
__NOTOC__ | __NOTOC__ | ||
| − | + | As requested at the Salt Lake City meeting, I have gathered a few images related to both segmentation and registration problems in radiation therapy. | |
| − | |||
| − | |||
| − | |||
| + | ===Interactive Segmentation=== | ||
| − | + | Rad Onc departments use interactive segmentation every day for both research and patient care. | |
| + | Prior to treatment planning, the target and critical structures are delineated in CT. The current state of the art is manual segmentation in axial view. A outline tool, used delineate the boundary, is generally prefered over a paintbrush tool that fills pixels. Commercial products generally support some subset of the following tools to assist the operator. | ||
| − | + | 1 contour interpolation between slices | |
| + | 2 boundary editing | ||
| + | 3 mixed axial/coronal/sagittal drawing | ||
| + | 4 livewire or intelligent scissors | ||
| + | 5 drawing constraints (e.g. constraints on volume overlap/distance) | ||
| + | 6 post-processing tools to nudge or smooth the boundary | ||
| − | + | ===Adaptive RT=== | |
| − | |||
| − | + | Below are some examples of anatomic change in head & neck and thorax. | |
| − | + | {| | |
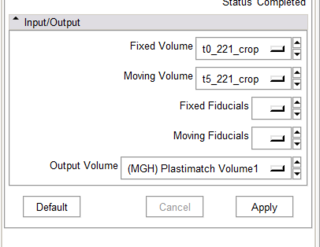
| − | + | |[[Image:gcsGUI.png|thumb|320px|Simple GUI for plastimatch]] | |
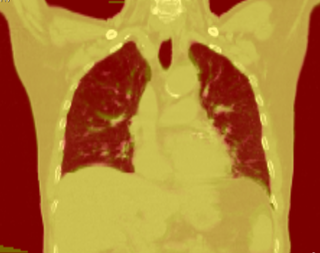
| − | + | |[[Image:gcsImage.jpg|thumb|320px|Registration output in Slicer]] | |
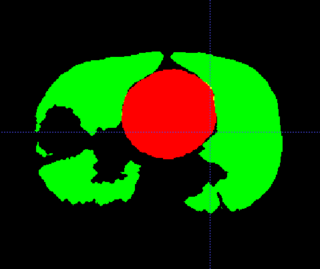
| − | + | |[[Image:gcsHeartLung.jpg|thumb|320px|Conversion of DICOM RT structures]] | |
| − | + | |} | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | ===General Discussion of Registration=== | |
| − | |||
| − | |||
| − | |||
| − | + | Deformable registration is still not as reliable as it should be. Image acquisition has residual artifacts which cause unrealistic deformations. Registration algorithms are not always robust, and require experimentation and tuning. Validation of registration results is not easy, since there are inadequate tools. Temporal regularization is generally not done, because of slow algorithms and large memory footprints. And so on. | |
| − | + | ===4D-CT Registration in Thorax=== | |
| − | + | Thorax is a special case. Patient images are acquired using 4D-CT, and radiation dose can computed for 3D volumes at each breathing phase. The volumes are aligned using deformable registration, and radiation dose is accumulated in a reference phase (e.g. exhale). Ideally this procedure is repeated to perform 4D treatment plan optimization. | |
| − | + | The sliding of the lungs against the chest wall is difficult to model. We sometimes segment the images at the pleural boundary. This allows us to separate the moving set of organs from the non-moving set, which are registered separately. Ideally we would always do this, but segmentation is manual and therefore we usually skip this step. | |
{| | {| | ||
| Line 52: | Line 41: | ||
|[[Image:gcsHeartLung.jpg|thumb|320px|Conversion of DICOM RT structures]] | |[[Image:gcsHeartLung.jpg|thumb|320px|Conversion of DICOM RT structures]] | ||
|} | |} | ||
| + | |||
| + | If you ignore the pleural boundary, registration of 4D-CT is considered "easy". | ||
| + | |||
| + | 1. Single-session imaging, so patient is already co-registered | ||
| + | 2. Single-session imaging, so no anatomic change | ||
| + | 3. High contrast of vessels against lung parenchema | ||
| + | |||
| + | State of the art is probably around 2-3 mm RMS error for point landmarks. | ||
<div style="margin: 20px;"> | <div style="margin: 20px;"> | ||
Revision as of 18:54, 29 January 2009
Home < NA-MIC < Projects < Collaboration < MGH RadOnc
As requested at the Salt Lake City meeting, I have gathered a few images related to both segmentation and registration problems in radiation therapy.
Interactive Segmentation
Rad Onc departments use interactive segmentation every day for both research and patient care. Prior to treatment planning, the target and critical structures are delineated in CT. The current state of the art is manual segmentation in axial view. A outline tool, used delineate the boundary, is generally prefered over a paintbrush tool that fills pixels. Commercial products generally support some subset of the following tools to assist the operator.
1 contour interpolation between slices 2 boundary editing 3 mixed axial/coronal/sagittal drawing 4 livewire or intelligent scissors 5 drawing constraints (e.g. constraints on volume overlap/distance) 6 post-processing tools to nudge or smooth the boundary
Adaptive RT
Below are some examples of anatomic change in head & neck and thorax.
General Discussion of Registration
Deformable registration is still not as reliable as it should be. Image acquisition has residual artifacts which cause unrealistic deformations. Registration algorithms are not always robust, and require experimentation and tuning. Validation of registration results is not easy, since there are inadequate tools. Temporal regularization is generally not done, because of slow algorithms and large memory footprints. And so on.
4D-CT Registration in Thorax
Thorax is a special case. Patient images are acquired using 4D-CT, and radiation dose can computed for 3D volumes at each breathing phase. The volumes are aligned using deformable registration, and radiation dose is accumulated in a reference phase (e.g. exhale). Ideally this procedure is repeated to perform 4D treatment plan optimization.
The sliding of the lungs against the chest wall is difficult to model. We sometimes segment the images at the pleural boundary. This allows us to separate the moving set of organs from the non-moving set, which are registered separately. Ideally we would always do this, but segmentation is manual and therefore we usually skip this step.
If you ignore the pleural boundary, registration of 4D-CT is considered "easy".
1. Single-session imaging, so patient is already co-registered 2. Single-session imaging, so no anatomic change 3. High contrast of vessels against lung parenchema
State of the art is probably around 2-3 mm RMS error for point landmarks.
Progress
- Working CLP program
- Got fiducials from Slicer -- Wow!
- Preliminary CTest interface
- DicomRT contour conversion
Todo
- Convert from CLP to scriptable or loadable module
- Improved visualization of registration output
- Improved interactivity of fiducials
- Export of deformed contours to DicomRT
References
- GC Sharp, Z Wu, N Kandasamy, "A Data Structure for B-Spline Registration," AAPM 50, Houston TX, July 2008.
- V Boldea, GC Sharp, SB Jiang, D Sarrut, “4D-CT lung motion estimation with deformable registration: quantification of motion nonlinearity and hysteresis,” Medical Physics, Vol 35, No 3, pp 1008-1018, March 2008.
- Z Wu, E Rietzel, V Boldea, D Sarrut, GC Sharp, "Evaluation of deformable registration of patient lung 4DCT with sub-anatomical region segmentations," Medical Physics, Vol 35, No 2, pp 775-81, February 2008.
- GC Sharp, N Kandasamy, H Singh, M Folkert, "GPU-based streaming architectures for fast cone-beam CT image reconstruction and demons deformable registration,” Physics in Medicine and Biology, Vol 52, No 19, pp 5771--83, October 7, 2007.