NA-MIC/Projects/Collaboration/MGH RadOnc
From NAMIC Wiki
Home < NA-MIC < Projects < Collaboration < MGH RadOnc
 Return to Project Week Main Page |
Key Investigators
- MGH: Greg Sharp, Marta Peroni
- NA-MIC: Steve Pieper, Wendy Plesniak
- BWH: Nicole Aucoin (fiducials), Katie Hayes (CTest)
Objective
The long term objective is to improve interactive tools for radiation therapy planning. Specifically, we would like to improve the reliability and usability of image registration and segmentation of critical organs.
Approach, Plan
Workshop goals
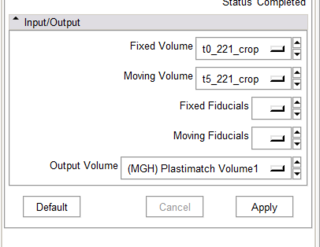
- Complete the CLP interface between our in-house registration software (plastimatch) and slicer3
- Get ideas how the user interface can be implemented
- Learn how to interface with slicer3 fiducial and label maps for interactive deformable registration
- Learn how CTest can be used for automated testing
Progress
Since last year
- Plastimatch software released under open source software license (BSD-style).
- The scaffolding code for slicer3 CLP is implemented, but not working because plastimatch doesn't read nrrd (yet).
Progress
Under construction.
 Return to Project Week Main Page |
References
- GC Sharp, Z Wu, N Kandasamy, "A Data Structure for B-Spline Registration," AAPM 50, Houston TX, July 2008.
- V Boldea, GC Sharp, SB Jiang, D Sarrut, “4D-CT lung motion estimation with deformable registration: quantification of motion nonlinearity and hysteresis,” Medical Physics, Vol 35, No 3, pp 1008-1018, March 2008.
- Z Wu, E Rietzel, V Boldea, D Sarrut, GC Sharp, "Evaluation of deformable registration of patient lung 4DCT with sub-anatomical region segmentations," Medical Physics, Vol 35, No 2, pp 775-81, February 2008.
- GC Sharp, N Kandasamy, H Singh, M Folkert, "GPU-based streaming architectures for fast cone-beam CT image reconstruction and demons deformable registration,” Physics in Medicine and Biology, Vol 52, No 19, pp 5771--83, October 7, 2007.