Difference between revisions of "NA-MIC/Projects/Collaboration/SBIA UPenn"
| (23 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| + | == '''DTI and HARDI analysis tools''' == | ||
| + | |||
| + | {| | ||
| + | |[[Image:ProjectWeek-2008.png|thumb|360px|Return to [[2008_Summer_Project_Week|Project Week Main Page]] ]] | ||
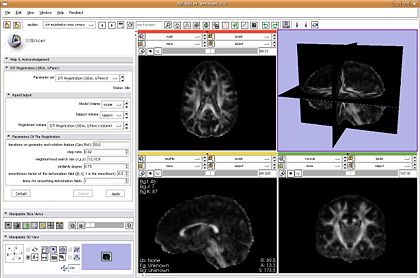
| + | |[[Image:DTIReg.JPG|thumb|420px|The non-linear DTI registration plugin.]] | ||
| + | |} | ||
{| | {| | ||
| − | |[[Image: | + | |[[Image:Registration_example.png|thumb|420px|Example of non-linear DTI registration: Row 1 is the template and row 2 is the average of 10 brains after deformable registration. ]] |
|} | |} | ||
| − | |||
__NOTOC__ | __NOTOC__ | ||
| − | |||
===Key Investigators=== | ===Key Investigators=== | ||
| − | *Ragini Verma, | + | *Ragini Verma, SBIA, UPenn <Ragini.Verma@uphs.upenn.edu> |
| − | + | *Christos Davatzikos <Christos.Davatzikos@uphs.upenn.edu> | |
| + | *Yang Li <Yang.Li@uphs.upenn.edu> (DTI integration) | ||
| + | *Luke Bloy <lbloy@seas.upenn.edu> (HARDI integration) | ||
| + | *Jinzhong Yang | ||
| + | *Dinggang Shen | ||
| + | *Carl-Fredrik Westin, BWH | ||
| + | *Luis Ibanez, Slicer | ||
| + | *Steve Pieper, Slicer | ||
| Line 16: | Line 27: | ||
<h1>Objective</h1> | <h1>Objective</h1> | ||
| − | + | To incorporate into Slicer, processing and analysis methods for DTI and HARDI being developed at the Section of Biomedical Image Analysis (SBIA), UPenn [http://www.rad.upenn.edu/sbia]. The main components will be plugins for DTI registration and manifold-based statistics followed by methods for HARDI registration and statistics. | |
| − | |||
</div> | </div> | ||
| Line 24: | Line 34: | ||
<h1>Approach, Plan</h1> | <h1>Approach, Plan</h1> | ||
| − | + | *'''DTI registration''': has been developed in SBIA over a period of 2 years, mainly using in-house code (not in ITK). This will be incorporated as a plugin. | |
| + | *'''DTI Statistics''': SPM-like package for statistics on the full tensor. The proposed format is again a plugin as it is based on code already existing in SBIA in C, C++. | ||
| + | *'''HARDI registration''': This is being developed in ITK, building on HARDI representation format currently under development | ||
| + | *'''HARDI statistics''': being developed in ITK | ||
</div> | </div> | ||
| Line 31: | Line 44: | ||
<h1>Progress</h1> | <h1>Progress</h1> | ||
| + | *DTI Registration: plugin is ready and was demonstrated | ||
| + | Two step process: | ||
| + | |||
| + | 1. Executable will be provided immediately (in discussion with : Steve Pieper) | ||
| + | |||
| + | 2. Source code will be checked in after some licensing issues (as it involves some legacy code) are resolved (in discussion with : Luis Ibanez) | ||
| + | |||
| + | * Tensor Demons registration code was found from Slicer 2.6 | ||
| + | 1. Luis Ibanez will be looking at reviving it in discussion with C-F Westin. | ||
| + | 2. We will help from our end if recoding is necessary. | ||
| + | *ODF estimation code is ready and will be checked into Namic Sandbox after preliminary testing. This will hopefully serve as the backend to Demian Wasserman's visulaization. | ||
</div> | </div> | ||
| Line 38: | Line 62: | ||
</div> | </div> | ||
| + | |||
| + | ===References=== | ||
| + | This work was supported in part by NIH grants R01-MH-079938 and R01-MH-070365. | ||
| + | |||
| + | * Link to [http://www.rad.upenn.edu/sbia SBIA web site: ] | ||
| + | * J. Yang, D. Shen, C. Davatzikos and R. Verma, Diffusion Tensor Image Registration Using Tensor Geometry and Orientation Features, in MICCAI 2008, New York. | ||
| + | * J. Yang, D. Shen, C. Davatzikos and R. Verma, Spatial Normalization of Diffusion Tensor Images Based on Anisotropic Segmentation, in SPIE Medical Imaging 2008, San Diego. | ||
| + | * R. Verma, P. Khurd, C. Davatzikos, [https://www.rad.upenn.edu/sbia/papers/181.pdf On Analyzing Diffusion Tensor Images by Identifying Manifold Structure using Isomaps], IEEE Transactions on Medical Imaging, 772-778, Vol. 26, No. 6, 2007 | ||
| + | * P. Khurd, R. Verma and C. Davatzikos, [https://www.rad.upenn.edu/sbia/papers/177.pdf Kernel-based Manifold Learning for Statistical Analysis of Diffusion Tensor Images], Information Processing in Medical Imaging (IPMI), 581-593, Vol. 4584, 2007 | ||
Latest revision as of 01:00, 27 June 2008
Home < NA-MIC < Projects < Collaboration < SBIA UPennDTI and HARDI analysis tools
 Return to Project Week Main Page |
Key Investigators
- Ragini Verma, SBIA, UPenn <Ragini.Verma@uphs.upenn.edu>
- Christos Davatzikos <Christos.Davatzikos@uphs.upenn.edu>
- Yang Li <Yang.Li@uphs.upenn.edu> (DTI integration)
- Luke Bloy <lbloy@seas.upenn.edu> (HARDI integration)
- Jinzhong Yang
- Dinggang Shen
- Carl-Fredrik Westin, BWH
- Luis Ibanez, Slicer
- Steve Pieper, Slicer
Objective
To incorporate into Slicer, processing and analysis methods for DTI and HARDI being developed at the Section of Biomedical Image Analysis (SBIA), UPenn [1]. The main components will be plugins for DTI registration and manifold-based statistics followed by methods for HARDI registration and statistics.
Approach, Plan
- DTI registration: has been developed in SBIA over a period of 2 years, mainly using in-house code (not in ITK). This will be incorporated as a plugin.
- DTI Statistics: SPM-like package for statistics on the full tensor. The proposed format is again a plugin as it is based on code already existing in SBIA in C, C++.
- HARDI registration: This is being developed in ITK, building on HARDI representation format currently under development
- HARDI statistics: being developed in ITK
Progress
- DTI Registration: plugin is ready and was demonstrated
Two step process:
1. Executable will be provided immediately (in discussion with : Steve Pieper)
2. Source code will be checked in after some licensing issues (as it involves some legacy code) are resolved (in discussion with : Luis Ibanez)
- Tensor Demons registration code was found from Slicer 2.6
1. Luis Ibanez will be looking at reviving it in discussion with C-F Westin.
2. We will help from our end if recoding is necessary.
- ODF estimation code is ready and will be checked into Namic Sandbox after preliminary testing. This will hopefully serve as the backend to Demian Wasserman's visulaization.
References
This work was supported in part by NIH grants R01-MH-079938 and R01-MH-070365.
- Link to SBIA web site:
- J. Yang, D. Shen, C. Davatzikos and R. Verma, Diffusion Tensor Image Registration Using Tensor Geometry and Orientation Features, in MICCAI 2008, New York.
- J. Yang, D. Shen, C. Davatzikos and R. Verma, Spatial Normalization of Diffusion Tensor Images Based on Anisotropic Segmentation, in SPIE Medical Imaging 2008, San Diego.
- R. Verma, P. Khurd, C. Davatzikos, On Analyzing Diffusion Tensor Images by Identifying Manifold Structure using Isomaps, IEEE Transactions on Medical Imaging, 772-778, Vol. 26, No. 6, 2007
- P. Khurd, R. Verma and C. Davatzikos, Kernel-based Manifold Learning for Statistical Analysis of Diffusion Tensor Images, Information Processing in Medical Imaging (IPMI), 581-593, Vol. 4584, 2007