Difference between revisions of "NA-MIC/Projects/External Collaboration/Measuring Alcohol and Stress Interaction"
From NAMIC Wiki
| Line 1: | Line 1: | ||
{| | {| | ||
|[[Image:ProjectWeek-2008.png|thumb|320px|Return to [[2008_Summer_Project_Week|Project Week Main Page]] ]] | |[[Image:ProjectWeek-2008.png|thumb|320px|Return to [[2008_Summer_Project_Week|Project Week Main Page]] ]] | ||
| − | + | ||
|} | |} | ||
| Line 12: | Line 12: | ||
* BWH: Kilian Pohl | * BWH: Kilian Pohl | ||
* Virginia Tech: Chris Wyatt, Vidya Rajagopalan | * Virginia Tech: Chris Wyatt, Vidya Rajagopalan | ||
| + | * Kitware: Luis Ibanez | ||
<div style="margin: 20px;"> | <div style="margin: 20px;"> | ||
| Line 40: | Line 41: | ||
# Study data on 8 subjects at two time points (alcohol naive and post-induction) organized | # Study data on 8 subjects at two time points (alcohol naive and post-induction) organized | ||
# GM/WM/CSF atlas will be constructed for the study data. | # GM/WM/CSF atlas will be constructed for the study data. | ||
| − | |||
| + | During Project Week | ||
| + | |||
| + | |||
| + | |||
| + | 1 Optimized segmentation parameters for vervet data | ||
| + | 2 Improved accuracy of subcortical segmentation in rhesus data set | ||
| + | |||
| + | {| | ||
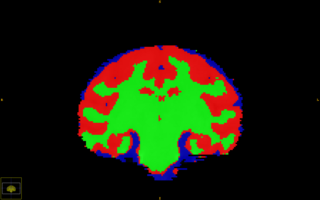
| + | |[[Image:Vervet_CorScrnShot.jpg|thumb|320px|Segmentation of major tissue classes in for vervet subject ]] | ||
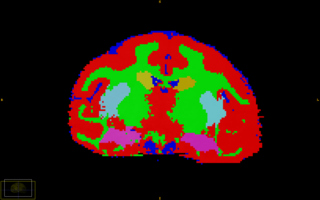
| + | |[[Image:Rhesus_HpcCorScrnShot.jpg|thumb|320px|Subcortical Segmentation of Rhesus subject]] | ||
| + | |} | ||
| + | |||
| + | 3 Developed an ITK utility for multiresolution demons registration. (Source code available for download from the NAMIC sandbox) | ||
| + | {| | ||
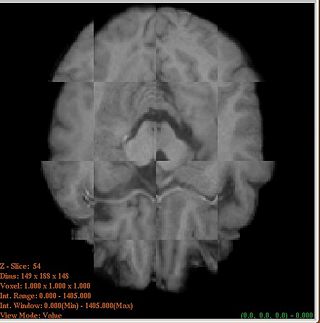
| + | |[[Image:CheckerB4Reg_Screenshot.jpg |thumb|320px|Checker Image of source and target before registration ]] | ||
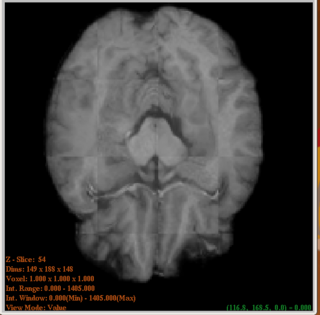
| + | |[[Image:CheckerAfterreg_Screenshot.png|thumb|320px|Checker Image of source and target after registration]] | ||
| + | |} | ||
Revision as of 21:51, 26 June 2008
Home < NA-MIC < Projects < External Collaboration < Measuring Alcohol and Stress Interaction Return to Project Week Main Page |
Key Investigators
- Wake Forest: Bob Kraft, Jim Daunais
- BWH: Kilian Pohl
- Virginia Tech: Chris Wyatt, Vidya Rajagopalan
- Kitware: Luis Ibanez
Objective
The objectives of this project are to:
- Optimize the EM Brain Classifier in Slicer on MRI images of vervets.
- Segment data from N subjects.
- Optimize GM/WM/CSF segmentation so that calculation of GM/WM volume and ratios between the two states can be automated.
- Optimize hippocampus, putamen and caudate segmentation so that volume comparison between states can be performed.
Approach, Plan
We will use the current implementation of the EM Segmenter in Slicer. Using a manual segmentation as an estimate of ground truth in a single subject, we will optimize the EM segmenter parameters over the classification error and running time.
Progress
Prior to the project week:
- Study data on 8 subjects at two time points (alcohol naive and post-induction) organized
- GM/WM/CSF atlas will be constructed for the study data.
During Project Week
1 Optimized segmentation parameters for vervet data 2 Improved accuracy of subcortical segmentation in rhesus data set
3 Developed an ITK utility for multiresolution demons registration. (Source code available for download from the NAMIC sandbox)