Difference between revisions of "Plug-In 3D Viewer based on XIP"
MelanieGrebe (talk | contribs) (Created page with '__NOTOC__ <gallery> Image:PW2009-v3.png|Project Week Main Page Image:genuFAp.jpg|Scatter plot of the original FA data through the genu of the corpus ...') |
MelanieGrebe (talk | contribs) |
||
| (12 intermediate revisions by 3 users not shown) | |||
| Line 2: | Line 2: | ||
<gallery> | <gallery> | ||
Image:PW2009-v3.png|[[2009_Summer_Project_Week|Project Week Main Page]] | Image:PW2009-v3.png|[[2009_Summer_Project_Week|Project Week Main Page]] | ||
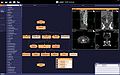
| − | Image: | + | Image:Xipexample.jpg|[http://openxip.org/ XIP: eXtensible Imaging Platform] |
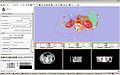
| − | Image: | + | Image:SlicerImage_lut_1.jpg|Volume Rendering in Slicer using XIP |
</gallery> | </gallery> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
==Key Investigators== | ==Key Investigators== | ||
| − | * | + | * Siemens Corporate Research: Lining Yang, Melanie Grebe |
| − | * | + | * Steve Pieper |
<div style="margin: 20px;"> | <div style="margin: 20px;"> | ||
| Line 21: | Line 14: | ||
<h3>Our Objective</h3> | <h3>Our Objective</h3> | ||
| − | + | Our objective is to create a new 3D Slicer Plug-in based on NCI's eXtensible Imaging Platform (XIP), which can then be used as a starting point for future projects involving NA-MIC and caBIG. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</div> | </div> | ||
| Line 32: | Line 20: | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
| − | <h3>Our Approach | + | <h3>Our Approach and Plan</h3> |
| − | |||
| − | |||
| − | + | We will implement a Host/Plug-in interface API that leverages the use of XIP modules (based on Open Inventor), and as a first example we will implement a simple 3D Viewer that can load and display DICOM data and visualize/navigate it using 3 synchronized MPR windows and a 3D volume rendering window, with the ability to render fused MR brain data. | |
</div> | </div> | ||
| Line 43: | Line 29: | ||
<h3>Our Progress</h3> | <h3>Our Progress</h3> | ||
| − | + | We implemented a prototype that directly embedded the XIP in the existing Volume Rendering Module of the 3D Slicer where we take the output volume and other parameters from Slicer/VTK and use them to build and render an XIP scenegraph using XIP's volume rendering nodes. | |
| + | |||
| + | Future work: | ||
| + | During the week we talked to Ron, Steve, Jim and many others to explore other ways of integrating the XIP and 3D Slicer. Among them are: (1) Uses the socket connection / Slicer Daemon to establish a host/plug-in architecture where we can leverage the strength of both the XIP/slicer world. One example is that XIP serves as a Dicom explorer host and slicer as a post-processing plugin. (2) Uses XIP to help the users to further process / perform visual programming of for example the slicer command line module results. | ||
| Line 52: | Line 41: | ||
==References== | ==References== | ||
| − | |||
| − | |||
| − | |||
| − | |||
| + | * [http://wiki.na-mic.org/Wiki/index.php/2009_Winter_Project_Week_Interactive_3D_Widgets_In_Slicer3 2009 Winter project week] | ||
</div> | </div> | ||
Latest revision as of 13:55, 25 June 2009
Home < Plug-In 3D Viewer based on XIPKey Investigators
- Siemens Corporate Research: Lining Yang, Melanie Grebe
- Steve Pieper
Our Objective
Our objective is to create a new 3D Slicer Plug-in based on NCI's eXtensible Imaging Platform (XIP), which can then be used as a starting point for future projects involving NA-MIC and caBIG.
Our Approach and Plan
We will implement a Host/Plug-in interface API that leverages the use of XIP modules (based on Open Inventor), and as a first example we will implement a simple 3D Viewer that can load and display DICOM data and visualize/navigate it using 3 synchronized MPR windows and a 3D volume rendering window, with the ability to render fused MR brain data.
Our Progress
We implemented a prototype that directly embedded the XIP in the existing Volume Rendering Module of the 3D Slicer where we take the output volume and other parameters from Slicer/VTK and use them to build and render an XIP scenegraph using XIP's volume rendering nodes.
Future work: During the week we talked to Ron, Steve, Jim and many others to explore other ways of integrating the XIP and 3D Slicer. Among them are: (1) Uses the socket connection / Slicer Daemon to establish a host/plug-in architecture where we can leverage the strength of both the XIP/slicer world. One example is that XIP serves as a Dicom explorer host and slicer as a post-processing plugin. (2) Uses XIP to help the users to further process / perform visual programming of for example the slicer command line module results.