Difference between revisions of "Project Week 25/Breast Segmentation in DCE-MRI via Deep Learning Approaches"
(Created page with "__NOTOC__ Back to Projects List ==Key Investigators== <!-- Key Investigator bullet points --> *Investigator 1 (Affiliation) *Investigator 2 (Af...") |
|||
| Line 3: | Line 3: | ||
Back to [[Project_Week_25#Projects|Projects List]] | Back to [[Project_Week_25#Projects|Projects List]] | ||
| + | '''Breast Cancer Analysis in DCE-MRI''' | ||
==Key Investigators== | ==Key Investigators== | ||
<!-- Key Investigator bullet points --> | <!-- Key Investigator bullet points --> | ||
| − | * | + | *Gabriele Piantadosi (University Federico II di Napoli, Italy)\ |
| − | + | ||
| − | + | ||
| + | ==Background== | ||
| + | In recent years Dynamic Contrast Enhanced-Magnetic Resonance Imaging (DCE-MRI) has gained popularity as an important complementary diagnostic methodology for early detection of breast cancer. It has demonstrated a great potential in the screening of high-risk women, in staging newly diagnosed breast cancer patients and in assessing therapy effects thanks to its minimal invasiveness and to the possibility to visualise 3D high resolution dynamic (functional) information not available with conventional RX imaging. | ||
| + | Among the major issues in developing CAD systems for breast DCE-MRI there are: (a) the detection of the suspicious region of interests (ROIs) as sensibly as possible, while simultaneously minimising the number of false alarms and (b) the classification of each segmented ROI according to its aggressiveness. This task is made harder by the peculiarity of DCE-MRI breast examinations: breast movements due to inspiration, a huge diversity of lesion types. | ||
==Project Description== | ==Project Description== | ||
| + | Segmentation of the breast parenchyma could be approached as a classification problem. | ||
| + | |||
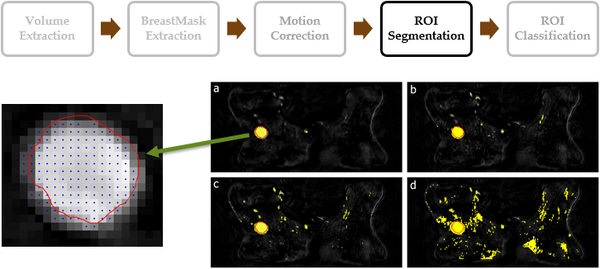
| + | [[File:Lesion Segmentation in DCE-MRI.png|thumb|center|upright=2.0|Lesion Segmentation in DCE-MRI]] | ||
| + | |||
| + | In the image, my previous proposal<ref>Marrone, S., Piantadosi, G., Fusco, R., Petrillo, A., Sansone, M., & Sansone, C. (2013, September). Automatic lesion detection in breast DCE-MRI. In International Conference on Image Analysis and Processing (pp. 359-368). Springer Berlin Heidelberg</ref>: classical machine learning approaches using Support Vector Machine (SVM) trained with dynamic features, extracted from a suitable pre-selected area by using a pixel-based approach. A pre-selection mask strongly improved the final result. | ||
| + | |||
| + | The novel proposed lesion detection module performs the segmentation of lesions in Regions of Interest (ROIs) by means of classification at a pixel level or as dense regions relying on deep approaches such as those proposed in<ref>Long, J., Shelhamer, E., & Darrell, T. (2015). Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (pp. 3431-3440)</ref>. | ||
| + | |||
{| class="wikitable" | {| class="wikitable" | ||
! style="text-align: left; width:27%" | Objective | ! style="text-align: left; width:27%" | Objective | ||
| Line 18: | Line 30: | ||
|- style="vertical-align:top;" | |- style="vertical-align:top;" | ||
|<!-- Objective bullet points --> | |<!-- Objective bullet points --> | ||
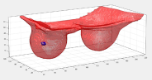
| + | *Lesion segmentation of 4D DCE-MRI volumes via deep approaches. | ||
| + | [[File:Segmented lesion in a 3D reconstructed Breast.png|frame|center|Segmented lesion in a 3D reconstructed Breast]] | ||
| + | *Results evaluation with 42 patients provided of manually segmented ground-truth. | ||
|<!-- Approach and Plan bullet points --> | |<!-- Approach and Plan bullet points --> | ||
| + | #Prepare the TensorFlow environment | ||
| + | #Prepare the Data | ||
| + | #Test the classical deep networks architectures | ||
| + | #Evaluate results in a k-fold flavour | ||
| + | #Try to do better by using a specifically designed network architecture. | ||
|<!-- Progress and Next steps (fill out at the end of project week), bullet points --> | |<!-- Progress and Next steps (fill out at the end of project week), bullet points --> | ||
| − | + | Relying on the segmented regions of interest (ROIs) a lesion malignity assessment via deep approaches should be performed. | |
|} | |} | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
==Background and References== | ==Background and References== | ||
| − | < | + | <references /> |
Revision as of 07:58, 24 June 2017
Home < Project Week 25 < Breast Segmentation in DCE-MRI via Deep Learning Approaches
Back to Projects List
Breast Cancer Analysis in DCE-MRI
Key Investigators
- Gabriele Piantadosi (University Federico II di Napoli, Italy)\
Background
In recent years Dynamic Contrast Enhanced-Magnetic Resonance Imaging (DCE-MRI) has gained popularity as an important complementary diagnostic methodology for early detection of breast cancer. It has demonstrated a great potential in the screening of high-risk women, in staging newly diagnosed breast cancer patients and in assessing therapy effects thanks to its minimal invasiveness and to the possibility to visualise 3D high resolution dynamic (functional) information not available with conventional RX imaging. Among the major issues in developing CAD systems for breast DCE-MRI there are: (a) the detection of the suspicious region of interests (ROIs) as sensibly as possible, while simultaneously minimising the number of false alarms and (b) the classification of each segmented ROI according to its aggressiveness. This task is made harder by the peculiarity of DCE-MRI breast examinations: breast movements due to inspiration, a huge diversity of lesion types.
Project Description
Segmentation of the breast parenchyma could be approached as a classification problem.
In the image, my previous proposal[1]: classical machine learning approaches using Support Vector Machine (SVM) trained with dynamic features, extracted from a suitable pre-selected area by using a pixel-based approach. A pre-selection mask strongly improved the final result.
The novel proposed lesion detection module performs the segmentation of lesions in Regions of Interest (ROIs) by means of classification at a pixel level or as dense regions relying on deep approaches such as those proposed in[2].
| Objective | Approach and Plan | Progress and Next Steps |
|---|---|---|
|
|
Relying on the segmented regions of interest (ROIs) a lesion malignity assessment via deep approaches should be performed. |
Background and References
- ↑ Marrone, S., Piantadosi, G., Fusco, R., Petrillo, A., Sansone, M., & Sansone, C. (2013, September). Automatic lesion detection in breast DCE-MRI. In International Conference on Image Analysis and Processing (pp. 359-368). Springer Berlin Heidelberg
- ↑ Long, J., Shelhamer, E., & Darrell, T. (2015). Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (pp. 3431-3440)