Difference between revisions of "Projects:ARRA:SlicerEM"
| Line 27: | Line 27: | ||
=Progress= | =Progress= | ||
| − | * 8/ | + | * 9/3/10 |
| + | * 8/27/10 | ||
** Integrated Workflow manager with QtModule | ** Integrated Workflow manager with QtModule | ||
** Review Workflow manager API | ** Review Workflow manager API | ||
| Line 37: | Line 38: | ||
*** changing some stderr output to stdout | *** changing some stderr output to stdout | ||
** Updating EMSegmenter tutorial for Slicer 3.6 | ** Updating EMSegmenter tutorial for Slicer 3.6 | ||
| − | * 8/ | + | * 8/20/10 |
** Ported anatomical tree widget and list of input channel - The anatomical tree is generic enough and re-used. | ** Ported anatomical tree widget and list of input channel - The anatomical tree is generic enough and re-used. | ||
** Added [[Projects:ARRA:SlicerEM#Image_gallery|Image gallery]] | ** Added [[Projects:ARRA:SlicerEM#Image_gallery|Image gallery]] | ||
| − | * 8/ | + | * 8/13/10 |
** Meet with Kilian - review panels and widgets and talked about which features should be improved, changed or omitted | ** Meet with Kilian - review panels and widgets and talked about which features should be improved, changed or omitted | ||
| − | * 8/ | + | * 8/6/10 |
** Now possible to enable wrapping of QtModule (Gui, UItools) in Slicer through CTK/PythonQt | ** Now possible to enable wrapping of QtModule (Gui, UItools) in Slicer through CTK/PythonQt | ||
** Discuss how TCL script interacted with the KWWidgets workflow manager | ** Discuss how TCL script interacted with the KWWidgets workflow manager | ||
| − | * 7/ | + | * 7/30/10 |
** The port of the Workflow manager is initiated | ** The port of the Workflow manager is initiated | ||
** Created Qt static panel skeleton corresponding to the step of MRI human brain workflow | ** Created Qt static panel skeleton corresponding to the step of MRI human brain workflow | ||
** Ability to script task in python is something important. Re-prioritize the task. | ** Ability to script task in python is something important. Re-prioritize the task. | ||
** Discussed how dynamic panel could be ported. | ** Discussed how dynamic panel could be ported. | ||
| − | * 7/ | + | * 7/23/10 |
** Reviewed [[Projects:ARRA:SlicerEM:Developer#Milestones|milestone]] and list of [[Projects:ARRA:SlicerEM:Developer:QtWidgets|widgets to port]] | ** Reviewed [[Projects:ARRA:SlicerEM:Developer#Milestones|milestone]] and list of [[Projects:ARRA:SlicerEM:Developer:QtWidgets|widgets to port]] | ||
** Created initial directory structure | ** Created initial directory structure | ||
Revision as of 15:24, 29 August 2010
Home < Projects:ARRA:SlicerEMAim
The EMSegmenter is a state-of-the-art segmentation tool within 3D Slicer. User feedback has reported that clinicians are currently unable to tune the approach to their acquisition protocol, as the user interface is too complex. This proposal addresses this issue by redesigning the user interface, focusing on hiding the complexity of the underlying segmentation algorithm. If successful, this will enable clinicians to automatically segment their own medical scans, even if the corresponding acquisition protocol deviates from the default setting for which the EMSegmenter is optimized.
Research Plan
The EMSegmenter is the result of 15 years of research in medical image segmentation. This has lead to a user interface that exposes a rich set of parameters. These parameters allow the tuning of the EMSegmenter to a wide variety of acquisition sequences. However, tuning these parameters is quite challanging. In addition, Slicer currently does not provide any tools for generating atlases, which are a set of parameters characterizing each structure of interest. We propose to address these issues by creating two user-friendly modules: one for generating the atlases and one for tuning the EMSegmenter to a specific acquisition sequence.
The first module, called Atlas Generator, builds the atlases characterizing each structure of interest. The user simply specifies the training data which can be done via querying XNAT, a database targeted towards medical image analysis. The user also selects the type of information to be extracted from the data. Possible types are the shape, intensity, or relative position of the structures of interest across the training set. Based on this input, the tool automatically generates the atlas.
The second module, called EMSegmenter-Simple, consists of a simple work flow that enables users to adjust the EMSegmenter to their specific acquisition sequence. As part of this proposal, we will create a library of templates, which parametrizes the tool to segmentation tasks frequently ancountered by our user community. User simply adopt the tool to their acquisition scenario by first selecting the proper template. In the second step, the user modifies the value of important parameters of the template. We simplify the tuning of these parameters by providing an instant feedback mechanism. The feedback mechanism updates the automatic segmentation according to the change in the parameter setting. This will allow users to to get an intuitive understanding about impact of certain parameters on the algorithm. In addition, each entry field will be accompanied with a help text. We will also create detailed documentation about the user interface and publish a tutorial for each template.
The project is viewed as successful if a properly trained clinician is able to modify the templates to their acquisition sequence within an hour.
Events
- Advanced EMSegmenter Training : How to parametrize the tool
- Where: 1249 Bolyston Street, Boston, MA
- When: 10 am - 1 pm , Feb 23, 2010
Key Personnel
10% Kilian Pohl
95% Dominique Belhachemi
5% Daniel Haehn
Kitware (Project Manager: Jean-Christophe Fillion-Robin)
Documentation
- EMSegmenter documentation for Slicer 3.6
- Developer page
Progress
- 9/3/10
- 8/27/10
- Integrated Workflow manager with QtModule
- Review Workflow manager API
- Fix bug related to PythonQt wrapping
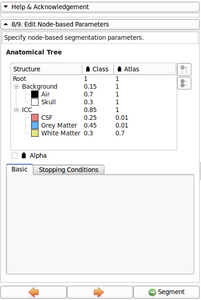
- Now possible to update atlas class weight, atlas weight and alpha value directly from the Anatomical tree
- Updated EMSegmenter related Wiki pages
- Testing EMSegmenter command line executable in a Sun Grid Engine (SGE) environment (branch 3.6 + trunk)
- working on related bugs 954, 955, 948
- changing some stderr output to stdout
- Updating EMSegmenter tutorial for Slicer 3.6
- 8/20/10
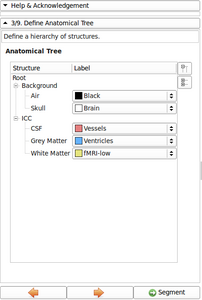
- Ported anatomical tree widget and list of input channel - The anatomical tree is generic enough and re-used.
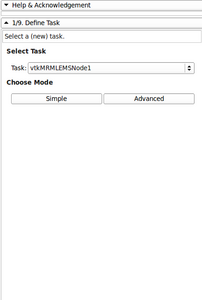
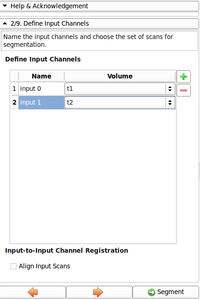
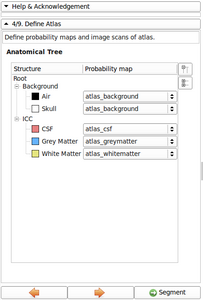
- Added Image gallery
- 8/13/10
- Meet with Kilian - review panels and widgets and talked about which features should be improved, changed or omitted
- 8/6/10
- Now possible to enable wrapping of QtModule (Gui, UItools) in Slicer through CTK/PythonQt
- Discuss how TCL script interacted with the KWWidgets workflow manager
- 7/30/10
- The port of the Workflow manager is initiated
- Created Qt static panel skeleton corresponding to the step of MRI human brain workflow
- Ability to script task in python is something important. Re-prioritize the task.
- Discussed how dynamic panel could be ported.
- 7/23/10
- Reviewed milestone and list of widgets to port
- Created initial directory structure
- Initiated the conversion of Graph/Algorithm/MRML code to separate shared module
- The port of existing TCL function related to processing is low priority
- 7/16/10
- First weekly meeting between Kitware and UPenn. Meeting will always be held after the Annotation/QT tcon.
- 7/9/10
- Remove bugs
- Set up contract with Kitware to port GUI to QT
- 7/2/10
- Hired Dominique Belhachemi
- Transfered grant from IBM to UPenn
- Wrote Progress Report
- 6/25/10
- Addressed all EMSegmenter bug in Mentis
- 6/18/10
- Revising task for MRI brain segmentation
- Started search for new hire
- Make EMSegmenter work with Slicer Superbuild
- 6/11/10
- Updated EMSegmenter documentation to reflect changes in Slicer 3.6 interface
- Fixed major bugs so that Slicer 3 does not crash when creating a new task
- 6/04/10
- Imbedded BRAINSFit registration in the pipeline
- 5/28/10
- Changed Simple version of EMSegmenter to work with new preprocessing structure
- 5/21/10
- Fixed class overview window
- 5/14/10
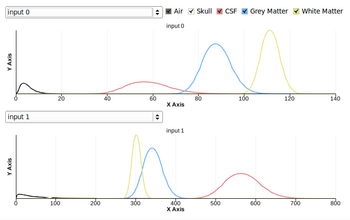
- Fixed Intensity graph distribution display
- 5/07/10
- Fixed EMSegmenter command line module
- 4/30/10
- Debugged EMSegmenter for Slicer 3.6 release
- 4/23/10
- Added segmentation button at each Step of the Wizard
- Integration of 3D/2D Bounding Box selection
- 4/16/10
- Updated Slicer 3.6 Documentation
- Modified workflow of Slicer 3.6
- 4/9/10
- Created simple interface for preprocessing
- 4/2/10
- Incorporated Bug fixes from Slicer 3.4 into trunk
- Removed bugs so that selecting tasks from a list properly works
- Rolled EMSegmenter back to Slicer version 3.4
- EMSegmenter Research environment now compiles under windows
- 3/26/10 Help users of EMSegment to adopt the tools to their needs
- 3/19/10 Manuscript was accepted by CVPR 2010 for poster presentation
- 3/12/10 Start organizing tutorials for EMSegmenter
- 3/05/10 Extended first step in EMSegmenter to include default task list
- 2/26/10 Removed bug so that api interface returns the same results as GUI version in Slicer2.6 . Trained members of CNL, BWH in segmenting lesions from MR brain images. Introduced CNL team to post processing tools for lesion segmentation.
- 2/19/10 Working with Andrey Fedorov, BWH, to customize the EMSegmenter to non-human primate brain images
- 2/12/10 Presented Slicer to IBM Health Care Division
- 2/05/10 Transferring Responsibilities for the EMSegmenter in Slicer 3.5 from MIT to IBM
- 1/29/10 Migrated EMSegmenter Research Environment from Slicer2 to Slicer3
- 1/22/10 Created slides for NAC Site visit
- 1/15/10 Started to resolve bugs that requires collaboration between Kitware, BWH, and IBM team
- 1/8/10 Trained members from BWH and Virginia Tech on using EMSegmenter on Non-Human primate data
- 1/1/10 Addressed all major bugs listed in Mantis for Slicer Version 3.4
- 12/25/09 Setup Slicer environment for new hire
- 12/18/09 Hired Yong Zhang to help with maintenance of EMSegmenter
- 12/11/09 Learned about programming user interfaces in Qt
- 12/04/09 Interviewed applicant for Post Doc position to work on this project
- 11/29/09 Removed bugs related to updated CMake version
- 11/20/09 Organize bugs related to EMSegmenter
- 11/13/09 Tcon featuring the EMSegmenter - participants Andriy Fedorov, Sylvain Jaume, Stuart Wallace, Kilian Pohl - Result of discussion
- Sylvain will fix EMSEgmenter bugs Slicer 3.5
- Kilian will fix any EMSegmenter bugs in Slicer 3.4 and earlier
- Andriy is developing a segmentation pipeline for the Wake Forest Data
- Stuart will do testing of the EMSegmenter module
- 11/06/09 Meet with Jean-Christophe Fillion-Robin from Kitware to discuss integration of Qt in 3D Slicer
- 10/30/09 Organized Monthly TCon between EMSegmenter developers
- 10/23/09 Organized onsite interview , got in contact with Steve Pieper to discuss next steps, installed Slicer3
- 10/17/09 Started interviewing postdoc as well as solving several HR issues for hiring personal
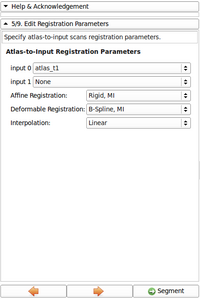
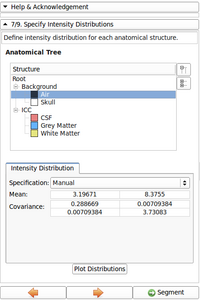
Image gallery
- EM Segmenter - Port to Qt