Difference between revisions of "Projects:RegistrationLibrary:RegLib C19"
(Created page with 'Back to ARRA main page <br> Back to Registration main page <br> [[Projects:RegistrationDocumentation:UseCaseInv…') |
m (Text replacement - "http://www.slicer.org/slicerWiki/index.php/" to "https://www.slicer.org/wiki/") |
||
| (22 intermediate revisions by one other user not shown) | |||
| Line 3: | Line 3: | ||
[[Projects:RegistrationDocumentation:UseCaseInventory|Back to Registration Use-case Inventory]] <br> | [[Projects:RegistrationDocumentation:UseCaseInventory|Back to Registration Use-case Inventory]] <br> | ||
| − | ==Slicer Registration Library | + | |
| − | {| style="color:#bbbbbb | + | == <small>v3.6.1</small> [[Image:Slicer3-6Announcement-v1.png|150px]] Slicer Registration Library Case #19: Multi-contrast group analysis: intra- and inter-subject registration of multi-contrast MRI == |
| − | |[[Image: | + | === Input === |
| − | |[[Image: | + | {| style="color:#bbbbbb; " cellpadding="2" cellspacing="0" border="0" |
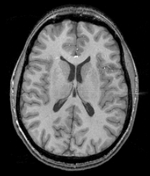
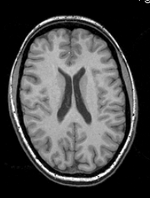
| − | + | |[[Image:RegLib_C19_thumb_N2_T1.png|150px|this is the main reference image. All images are aligned into this space]] | |
| − | |[[Image: | + | |[[Image:RegArrow_Affine.png|70px|lleft]] |
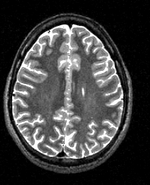
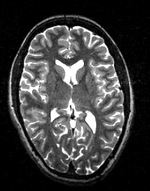
| − | |[[Image: | + | |[[Image:RegLib_C19_thumb_N2_T2.png|150px|this is the 1st intra-subject moving target, to be aligned with the main reference directly]] |
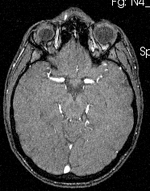
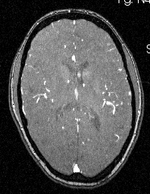
| − | | | + | |[[Image:RegLib_C19_thumb_N2_MRA.png|150px|this is the 2nd intra-subject moving target, to be aligned with the main reference directly]] |
| − | + | |- | |
| − | + | |subj 1: T1 | |
| − | + | | | |
| − | + | |subj 1: T2 | |
| + | |subj 1: MRA | ||
|- | |- | ||
| − | |[[Image: | + | |[[Image:RegArrow_NonRigidVert.png|70px|lleft]] |
| − | | | + | | |
| | | | ||
| − | |[[Image: | + | | |
| − | |[[Image: | + | |- |
| + | |[[Image:RegLib_C19_thumb_N4_T1.png|150px|this is both the fixed target for the 2nd subject and the moving image for inter-subject registration]] | ||
| + | |[[Image:RegArrow_Affine.png|70px|lleft]] | ||
| + | |[[Image:RegLib_C19_thumb_N4_T2.png|150px|this is a moving image to be aligned indirectly to the main ref]] | ||
| + | |[[Image:RegLib_C19_thumb_N4_MRA.png|150px|his is a moving image to be aligned indirectly to the main ref ]] | ||
|- | |- | ||
| − | | | + | |subj 2: T1 |
| − | |||
| | | | ||
| − | | | + | |subj 2: T2 |
| − | | | + | |subj 2: MRA |
|} | |} | ||
| + | |||
| + | *T1: 1mm isotropic; 176 x 256 x 176 | ||
| + | *T2: 1mm isotropic; 192 x 256 x 128 | ||
| + | *MRA: 0.51 x 0.51 x 0.8 mm; 448 x 448 x 128 | ||
| + | |||
| + | === Modules === | ||
| + | *'''Slicer 3.6.1 recommended modules: [https://www.slicer.org/wiki/Modules:BRAINSFit BrainsFit]''' | ||
| + | *For the inter-subject nonrigid portion: [https://www.slicer.org/wiki/Modules:DeformableB-SplineRegistration-Documentation-3.6 Fast nonrigid BSpline] | ||
| + | *For combining the transforms: copy and paste in a Text Editor | ||
| + | *For applying the transforms and resampling: [https://www.slicer.org/wiki/Modules:ResampleScalarVectorDWIVolume-Documentation-3.6 Resample Scalar/Vector/DWI Volume] | ||
===Objective / Background === | ===Objective / Background === | ||
| − | This is an example of inter-subject | + | This is an example of multi-stage registration containing both intra- and inter-subject alignment. We have two sets of multi-sequence MRI for two subjects, each comprised of a T1, T2 and perfusion MRA scan. We ant everything aligned to a single space to enable regional or voxel-based group comparison. |
| − | |||
=== Keywords === | === Keywords === | ||
| − | MRI, brain, head, inter-subject, | + | multi-stage registration, MRI, brain, head, multi-contrast, inter-subject, group analysis, MRA, T2 |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
=== Methods === | === Methods === | ||
| − | # | + | #align T2 and MRA of each subject to their respective T1 via an affine transform |
| − | + | #align the T1 of subject 2 to the T1 of subject 1 via a non-rigid transform, this establishes the inter-subject mapping | |
| − | + | #combine the affine transforms of subject 2 T2 and MRA with the above nonrigid and resample the two images into the reference space | |
| − | |||
| − | |||
| − | # | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | # | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
=== Registration Results=== | === Registration Results=== | ||
| − | + | [[Image:RegLib_C19_unreg.gif|300px|unregistered]] unregistered (left) | |
| − | |[[Image: | + | [[Image:RegLib_C19_reg.gif|300px|registered]] registered series : Affine + Fast BSpline (3x3x3). Note the animated gif cycles through both subjects and intra-subject scans. <br> |
| − | | | + | |
| + | [[Image:RegLib_C19_AGif_Intra_N2.gif|300px|registered]] registered intra-subject N2 [[Image:RegLib_C19_AGif_Intra_N4.gif|300px|registered]] registered intra-subject N4 (right): Affine only<br> | ||
| + | [[Image:RegLib_C19_AGif_N4-N2_T1.gif|300px|registered]] registered N4-N2 (Affine+BSpline BRAINSfit 7x7x7)<br> | ||
| + | [[Image:RegLib_C19_AGif_N4-N2_T2.gif|300px|registered]] registered N4-N2 (Affine+BSpline BRAINSfit 7x7x7)<br> | ||
| + | [[Image:RegLib_C19_AGif_N4-N2_MRA.gif|300px|registered]] registered N4-N2 (Affine+BSpline BRAINSfit 7x7x7)<br> | ||
| + | [[Image:RegLib_C19_AGif_BSplDef.gif|300px|registered]] registered N4-N2 BSpline deformation only applied to N4<br> | ||
===Download === | ===Download === | ||
| − | *[[Media: | + | *Data |
| − | * | + | **[[Media:RegLib_C19_DATA.zip|download '''image data'''<small> (Original Data, Result transforms, parameter presets, zip file 81 MB) </small>]] |
| − | *[[Media: | + | *Presets |
| − | * | + | **[[Media:RegLib_C19_RegPresets.mrml|download '''registration parameter presets''' <small> (mrml file, 12 kB) </small>]] |
| − | *[[Media: | + | *Documentation |
| − | + | **[[Media:RegLib_C19_Tutorial.ppt|download '''guided tutorial''' <small> (power point file, 1.5 MB) </small>]] | |
[[Projects:RegistrationDocumentation:ParameterPresetsTutorial|Link to User Guide: How to Load/Save Registration Parameter Presets]] | [[Projects:RegistrationDocumentation:ParameterPresetsTutorial|Link to User Guide: How to Load/Save Registration Parameter Presets]] | ||
=== Discussion: Registration Challenges === | === Discussion: Registration Challenges === | ||
| − | * | + | *We have two separate sets of registrations to combine. While theoretically possible to simply align every scan with the reference directly, this is likely to produce inferior results. Basic rule of thumb is to register to the image that is closest in anatomy and contrast, in that order. |
| − | *the | + | *the MRA has a strongly clipped FOV and low tissue contrast |
| − | *the | + | *Currently (Slicer 3.6) the concatenation of linear and nonlinear transforms is not (yet) supported directly in slicer. However this is an easy and fast procedure that can be done with any text editor. We merely copy the affine portion from one transform file and paste it into the nonrigid transform file. |
=== Discussion: Key Strategies === | === Discussion: Key Strategies === | ||
| − | + | #For the intra-subject affine portion: [https://www.slicer.org/wiki/Modules:BRAINSFit BRAINSfit] | |
| − | + | #For the inter-subject nonrigid portion: [https://www.slicer.org/wiki/Modules:DeformableB-SplineRegistration-Documentation-3.6 Fast nonrigid BSpline] | |
| + | #For combining the transforms: copy and paste in a Text Editor | ||
| + | #For applying the transforms and resampling: [https://www.slicer.org/wiki/Modules:ResampleScalarVectorDWIVolume-Documentation-3.6 Resample Scalar/Vector/DWI Volume] | ||
=== Acknowledgments === | === Acknowledgments === | ||
| − | *dataset provided by | + | *dataset provided by the [http://ij.itk.org/midas/community/view/21 UNC Midas Database of healthy volunteers] |
Latest revision as of 18:07, 10 July 2017
Home < Projects:RegistrationLibrary:RegLib C19Back to ARRA main page
Back to Registration main page
Back to Registration Use-case Inventory
v3.6.1  Slicer Registration Library Case #19: Multi-contrast group analysis: intra- and inter-subject registration of multi-contrast MRI
Slicer Registration Library Case #19: Multi-contrast group analysis: intra- and inter-subject registration of multi-contrast MRI
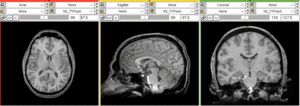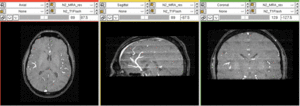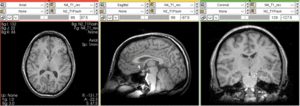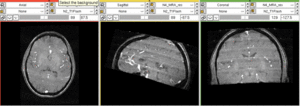
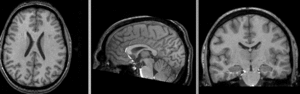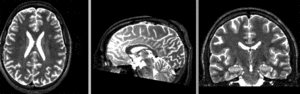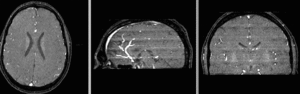
Input
|
|
|
|
| subj 1: T1 | subj 1: T2 | subj 1: MRA | |
|
|||
|

|
|
|
| subj 2: T1 | subj 2: T2 | subj 2: MRA |
- T1: 1mm isotropic; 176 x 256 x 176
- T2: 1mm isotropic; 192 x 256 x 128
- MRA: 0.51 x 0.51 x 0.8 mm; 448 x 448 x 128
Modules
- Slicer 3.6.1 recommended modules: BrainsFit
- For the inter-subject nonrigid portion: Fast nonrigid BSpline
- For combining the transforms: copy and paste in a Text Editor
- For applying the transforms and resampling: Resample Scalar/Vector/DWI Volume
Objective / Background
This is an example of multi-stage registration containing both intra- and inter-subject alignment. We have two sets of multi-sequence MRI for two subjects, each comprised of a T1, T2 and perfusion MRA scan. We ant everything aligned to a single space to enable regional or voxel-based group comparison.
Keywords
multi-stage registration, MRI, brain, head, multi-contrast, inter-subject, group analysis, MRA, T2
Methods
- align T2 and MRA of each subject to their respective T1 via an affine transform
- align the T1 of subject 2 to the T1 of subject 1 via a non-rigid transform, this establishes the inter-subject mapping
- combine the affine transforms of subject 2 T2 and MRA with the above nonrigid and resample the two images into the reference space
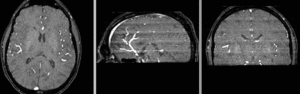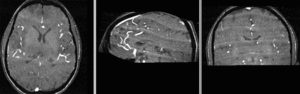
Registration Results
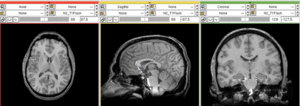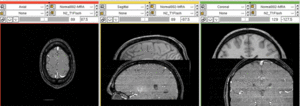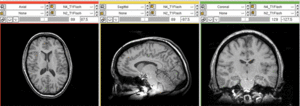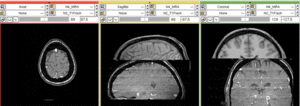 unregistered (left)
unregistered (left)
 registered series : Affine + Fast BSpline (3x3x3). Note the animated gif cycles through both subjects and intra-subject scans.
registered series : Affine + Fast BSpline (3x3x3). Note the animated gif cycles through both subjects and intra-subject scans.
 registered intra-subject N2
registered intra-subject N2 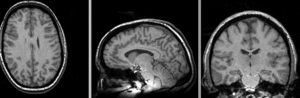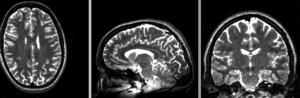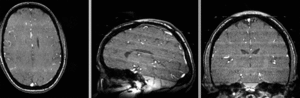 registered intra-subject N4 (right): Affine only
registered intra-subject N4 (right): Affine only
 registered N4-N2 (Affine+BSpline BRAINSfit 7x7x7)
registered N4-N2 (Affine+BSpline BRAINSfit 7x7x7)
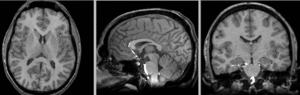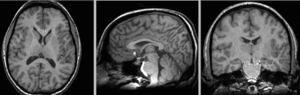
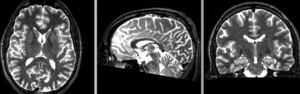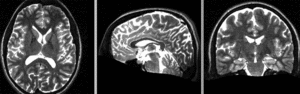 registered N4-N2 (Affine+BSpline BRAINSfit 7x7x7)
registered N4-N2 (Affine+BSpline BRAINSfit 7x7x7)
 registered N4-N2 (Affine+BSpline BRAINSfit 7x7x7)
registered N4-N2 (Affine+BSpline BRAINSfit 7x7x7)
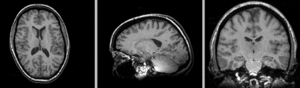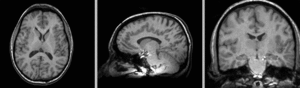 registered N4-N2 BSpline deformation only applied to N4
registered N4-N2 BSpline deformation only applied to N4
Download
- Data
- Presets
- Documentation
Link to User Guide: How to Load/Save Registration Parameter Presets
Discussion: Registration Challenges
- We have two separate sets of registrations to combine. While theoretically possible to simply align every scan with the reference directly, this is likely to produce inferior results. Basic rule of thumb is to register to the image that is closest in anatomy and contrast, in that order.
- the MRA has a strongly clipped FOV and low tissue contrast
- Currently (Slicer 3.6) the concatenation of linear and nonlinear transforms is not (yet) supported directly in slicer. However this is an easy and fast procedure that can be done with any text editor. We merely copy the affine portion from one transform file and paste it into the nonrigid transform file.
Discussion: Key Strategies
- For the intra-subject affine portion: BRAINSfit
- For the inter-subject nonrigid portion: Fast nonrigid BSpline
- For combining the transforms: copy and paste in a Text Editor
- For applying the transforms and resampling: Resample Scalar/Vector/DWI Volume
Acknowledgments
- dataset provided by the UNC Midas Database of healthy volunteers