Difference between revisions of "ProstateSegmentationAHM2009"
From NAMIC Wiki
| (4 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
{| | {| | ||
|[[Image:NAMIC-SLC.jpg|thumb|320px|Return to [[2009_Winter_Project_Week|Project Week Main Page]] ]] | |[[Image:NAMIC-SLC.jpg|thumb|320px|Return to [[2009_Winter_Project_Week|Project Week Main Page]] ]] | ||
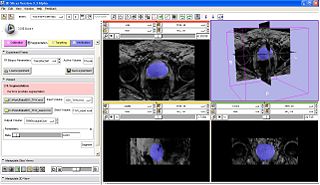
| − | |[[Image: | + | |[[Image:TRPB_ProstateSegmentation.JPG|Prostate segmentation integrated inside TR Prostate biopsy interactive module|thumb|320px]] |
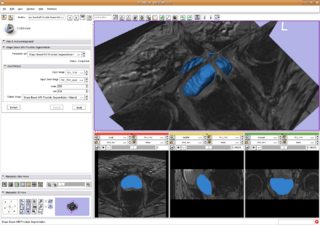
| + | |[[Image:ShapeBasePstSegSlicer.png|Shape base prostate segmentation in Slicer through command line module|thumb|320px]] | ||
|} | |} | ||
| Line 19: | Line 20: | ||
<h1>Objective</h1> | <h1>Objective</h1> | ||
| − | For the task of prostate segmentation, we provide two tools: 1. shape based and 2. semi-automatic random walk based. The goal is to put both algorithm into Slicer3 | + | For the task of prostate segmentation, we provide two tools: 1. shape based and 2. semi-automatic random walk based. The goal is to put both algorithm into Slicer3. |
</div> | </div> | ||
| Line 26: | Line 27: | ||
<h1>Approach, Plan</h1> | <h1>Approach, Plan</h1> | ||
| − | Algorithm 1. is a | + | Algorithm 1. is a Shape based segmentation. The shape of prostates are learned and then the new image is segmented using the shapes learned. Algorithm 2. is based on the Random Walks segmentation algorithm. It need more human input but the result could be interactively improved arbitrarily close to user's expectation. |
</div> | </div> | ||
| Line 34: | Line 35: | ||
<h1>Progress</h1> | <h1>Progress</h1> | ||
| − | + | During the project week, we achieved: | |
| − | |||
| − | |||
| − | |||
| − | |||
| + | * Integrated semi-auto algorithm (algorithm 2) into Trans Rectal Prostate biopsy interactive loadable module, shown in the middle figure above. | ||
| + | * Integrated shape based algorithm (algorithm 1) into Slicer3 as command line module, shown in the right figure above. | ||
</div> | </div> | ||
Latest revision as of 23:42, 8 January 2009
Home < ProstateSegmentationAHM2009 Return to Project Week Main Page |
Key Investigators
- Gabor Fichtinger, Purang Abolmaesumi, David Gobbi, Siddharth Vikal; Queen’s University
- Allen Tannenbaum, Yi Gao; Georgia Tech
- Katie Hayes, Brigham and Women's Hospital
Objective
For the task of prostate segmentation, we provide two tools: 1. shape based and 2. semi-automatic random walk based. The goal is to put both algorithm into Slicer3.
Approach, Plan
Algorithm 1. is a Shape based segmentation. The shape of prostates are learned and then the new image is segmented using the shapes learned. Algorithm 2. is based on the Random Walks segmentation algorithm. It need more human input but the result could be interactively improved arbitrarily close to user's expectation.
Progress
During the project week, we achieved:
- Integrated semi-auto algorithm (algorithm 2) into Trans Rectal Prostate biopsy interactive loadable module, shown in the middle figure above.
- Integrated shape based algorithm (algorithm 1) into Slicer3 as command line module, shown in the right figure above.