Difference between revisions of "Summer project week 2011 Workflows SOA"
| (10 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
<gallery> | <gallery> | ||
Image:PW-MIT2011.png|[[2011_Summer_Project_Week#Projects|Projects List]] | Image:PW-MIT2011.png|[[2011_Summer_Project_Week#Projects|Projects List]] | ||
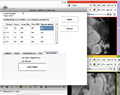
| − | Image: | + | Image:SlicerWF_LA_GUI.png|Left Atrium Ablation Follow-Up WF GUI |
</gallery> | </gallery> | ||
| − | ''' | + | '''Workflows and Service Oriented Architecture Modules for Slicer4 as Extensions''' |
==Key Investigators== | ==Key Investigators== | ||
| − | * Brigham and Womens Hospital, Harvard Medical School: Alexander Zaitsev, Wendy Plesniak, Ron | + | * Brigham and Womens Hospital, Harvard Medical School: Alexander Zaitsev, Wendy Plesniak, Charles Guttmann, Ron Kikinis |
| Line 16: | Line 16: | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
| − | #: | + | #:Thin clients for Slicer4 to support standardized workflows |
| − | #: | + | #: |
| − | #: ( | + | #:Automation of image processing steps, data management |
| + | #: | ||
| + | #:Combination of automated methods with human activities | ||
| + | #:Case Studies: Left Atrial Ablation follow-up, Brain PET/CT/MRI scans processing for cancer treatmen follow-up, brain atrophy estimation | ||
| + | #:Major methods/algorithms used by workflows: Registration (BRAINSFit, BRAINSResample), Biasfield correction | ||
</div> | </div> | ||
| Line 25: | Line 29: | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| − | #:Trying | + | #:Trying to build existing thin clients (Slicelets) |
| − | #: | + | #:as Slicer4 Extensions. |
| − | #: | + | #:Add Left Atrium Ablation scar segmentation into the CARMA workflow (RSS or LA Segmenter?) |
| + | #:Incorporate MS Lesion EM Segmentation Scenario into the workflows | ||
</div> | </div> | ||
| Line 34: | Line 39: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | + | '''Slicer4 Extensions and Custom Builds:''' | |
| − | < | + | <br> Learned how to do. Have to deal with package build for 32-bit Mac |
| − | + | <br> | |
| + | '''CARMA workflow''':<br> | ||
| + | Gave several Life demos (including) general SOA/workflows approach. Identified ergonomic to be addressed in CARMA WF GUI <br> | ||
| + | Segmented hyperintencity regions at left atrium wall (RSS module). Issue: overestimation, need to add more thresholding controls to RSS.<br> | ||
| + | '''MS Lesion Segmentation'''<br> | ||
| + | Worked with EM Segmenter module builders on adult and pediatric cases segmentation<br> | ||
| + | Lesion overestimation have to be addressed: can be done with manual correction<br> | ||
| + | Long running pre-processing steps make SOA/WF approach preferable for Lesion Segmentation <br> | ||
</div> | </div> | ||
</div> | </div> | ||
| + | |||
| + | <gallery> | ||
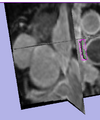
| + | Image:SlicerWF_LA_Sample.png|Manual Segmentation Sample(Utah Team) | ||
| + | Image:SlicerWF_LA_RSS.png|RSS Segmentation Attempt | ||
| + | </gallery> | ||
<div style="width: 97%; float: left;"> | <div style="width: 97%; float: left;"> | ||
| − | + | [[http://wiki.na-mic.org/Wiki/index.php/Winter_project_week_2011_Workflows_SOA Winter_project_week_2011_Workflows_SOA]] Winter 2001 Project Week Page | |
| − | + | <br> | |
| + | [[http://www.na-mic.org/Wiki/index.php/DBP3:Utah Left Atrium Ablation Project, CARMA Wokflow]] CARMA Workflow Page | ||
| + | |||
| + | [[http://wiki.na-mic.org/Wiki/index.php/DBP3:Utah:RegSegPipeline CARMA Wokflow Preprocessing Pipeline]] CARMA Wokflow Preprocessing Pipeline | ||
==Delivery Mechanism== | ==Delivery Mechanism== | ||
| Line 52: | Line 72: | ||
This work will be delivered to the NAMIC Kit as a | This work will be delivered to the NAMIC Kit as a | ||
| − | |||
| − | |||
##Built-in: | ##Built-in: | ||
##Extension -- commandline: | ##Extension -- commandline: | ||
##Extension -- loadable: | ##Extension -- loadable: | ||
Latest revision as of 19:33, 24 June 2011
Home < Summer project week 2011 Workflows SOAWorkflows and Service Oriented Architecture Modules for Slicer4 as Extensions
Key Investigators
- Brigham and Womens Hospital, Harvard Medical School: Alexander Zaitsev, Wendy Plesniak, Charles Guttmann, Ron Kikinis
Objective
- Thin clients for Slicer4 to support standardized workflows
- Automation of image processing steps, data management
- Combination of automated methods with human activities
- Case Studies: Left Atrial Ablation follow-up, Brain PET/CT/MRI scans processing for cancer treatmen follow-up, brain atrophy estimation
- Major methods/algorithms used by workflows: Registration (BRAINSFit, BRAINSResample), Biasfield correction
Approach, Plan
- Trying to build existing thin clients (Slicelets)
- as Slicer4 Extensions.
- Add Left Atrium Ablation scar segmentation into the CARMA workflow (RSS or LA Segmenter?)
- Incorporate MS Lesion EM Segmentation Scenario into the workflows
Progress
Slicer4 Extensions and Custom Builds:
Learned how to do. Have to deal with package build for 32-bit Mac
CARMA workflow:
Gave several Life demos (including) general SOA/workflows approach. Identified ergonomic to be addressed in CARMA WF GUI
Segmented hyperintencity regions at left atrium wall (RSS module). Issue: overestimation, need to add more thresholding controls to RSS.
MS Lesion Segmentation
Worked with EM Segmenter module builders on adult and pediatric cases segmentation
Lesion overestimation have to be addressed: can be done with manual correction
Long running pre-processing steps make SOA/WF approach preferable for Lesion Segmentation
[Winter_project_week_2011_Workflows_SOA] Winter 2001 Project Week Page
[Left Atrium Ablation Project, CARMA Wokflow] CARMA Workflow Page
[CARMA Wokflow Preprocessing Pipeline] CARMA Wokflow Preprocessing Pipeline
Delivery Mechanism
This work will be delivered to the NAMIC Kit as a
- Built-in:
- Extension -- commandline:
- Extension -- loadable: