Mbirn: MRI Diffusion Tools
UPDATES
March 7, 2006 T-Con notes (R Gollub)
In Attendence: S. Pieper, S. Mori, J. Fallon, R. Gollub, M. Panzenboeck, M. Perry
Progress:
- Susumu shared an interim analysis summarizing the 10 fiber tracking results from subjects 2, 3, and 4_2 and emailed us an updated table of results and manuscript. He noted that he had to go back to complete the QA of a few tracks that did not come out properly and we all agreed to allocate about an hour during the Sedona meeting to review the questionable areas and agree on an updated set of definitions with Susumu to lead us. We are very close to having final data for the manuscript.
- Agreed to wait until AHM to choose our next project. Much of the meeting is devoted to development of DTI plans and we will be part of it. All of us in this group will participate in those sessions.
Action Items:
- Susumu to finish his analysis and prepare for our hour session (Tuesday at 1:00 PM)
- Everyone to think about clinically relevant key issues in DTI analysis to contribute to the Sedona discussions
- Randy to contact the Pediatric Neuroimaging Initiative DTI expert (Carlo Pierpaoli) to get an update on what they have done. Done- in fact Carlo is going to join us for the Sedona meeting!
- Randy to find out if MGH is a Pediatric Neuroimaging Initiative acquisition site. Done- no the PI is Michael Rivkin at Children's Hosptial.
February 7, 2006 T-Con notes (R Gollub/K Helmer)
In Attendence (and all on time again, U all rule!) S. Pieper, S. Mori, J. Fallon, R. Gollub, M. Panzenboeck, A. Caprihan, K. Helmer. M. Perry could not attend due to a conflict with study data acquisition.
Progress:
- Susumu's further analysis of the fiber tracks from subject 4_1 shows that agreement looks pretty good. The fit is less good for corpus callosum. He asked all raters to send him the coronal slice number corresponding to the beginning of the track to see if he can figure out how to improve the specificity of the description for that track. All have done this except for Martina who will email this information Susumu. Susumu will revise the protocol to make this clearer.
- Michelle, Martina, Arvind completed the second round of data collection on time. Jim is still working on his.
Discussion of how to characterize the spatial characteristics of track overlap. Steve Pieper suggested that the STAPLE algorithm might be useful. This led to a discussion of what should be our next plans for M-BIRN work on DTI.
Discussion of what should be the next areas for the group to focus on. Some suggestions:
- How to update DTIStudio?
- How do different DTI tools respond when faced with the same data?
- How to register tensor data? What parameters might be used for the registration?
- What is the lower limit of tract size for DTI analysis?
The issue of understanding diffusion sequence parameters and how they are calculated (and stored) for different cases, such as oblique images is not well understood.
Can we come up with a table for the general user that makes software recomendations based on their data quality and what they are looking for?
Action plans:
- BY THIS FRIDAY FEBRUARY 10th: Michelle, Martina and Arvind (and Jim if he can finish his tracks by Friday) are to email zipped files of all 10 fiber tracking results from subjects 2, 3, and 4_2 to Arvind (arvindc@unm dot edu) who will run the matlab scripts to generate the reliability data.
- BY TUESDAY FEBRUARY 14th: Arvind will send these results to Susumu who will finish the analysis and incorporate the results into the manuscript
- We will meet once more by T-con to review these results on Tuesday February 28th at 11 AM EST/8 AM PST.
- See M-BIRN AHM schedule Mbirn:SpringConference2006 and email Randy or Karl feedback to improve planning
December 9, 2005 T-Con notes (R Gollub)
In Attendence (and all on time!) S. Pieper, S. Mori, J. Fallon, M. Perry, R. Gollub, M. Panzenboeck, A. Caprihan
Progress: All 3 sites completed data collection on time. Susumu's initial first pass analysis shows that all sent the correct data structures and agreement looks good. Good job team! The full analysis is not yet complete. The person from his lab who led this project moved away and is also out on maternity leave.
Action Plans:
- The raters (Michelle, Martina, Arvind, and Jim) are going to complete the exact same analysis on data from subjects 2, 3 and 4_2, obtaining both sets of tracks (see notes from 11/7/05 for exact instructions). Deadline for this is January 6th. Send CD to Susumu.
- Susumu will find another person in his lab group to help with this and in parallel will send his IDL code for performing the more detailed analysis of inter-rater reliabiity to Arvind and between these two efforts they will get the final analysis done. Target deadline for all analysis to be complete is January 20th, 2006.
- Some of us will attend the NA-MIC All Hands Meeting AHM2006_ValidationWorkshop. This discussion will help inform us of what direction to take after this project is complete.
- Our next T-con is Friday January 20th at 2 PM EST/11 AM PST
Steve Pieper and Jonathan Farrell are still working on the generation of NRRD readers for all the DTI datasets that Susumu has posted on his own website. Jim Fallon is keen to do some comparison across tractography tools using this data so is encouraging the team to finish this up!
Steve Pieper reported that BWH and affiliated researchers are interested in the topic of non-linear registration (multi-channel demon's algorithm) of subjects into a group atlas. Susumu confirmed that the current atlases are made with linear (affine) registration and that there's a possibility that a non-linear registration process might be desirable and is worth testing. Susumu also confirmed that 30 of the data sets being made available for the mBIRN testing will include the manually defined tracts and can be used for testing this non-linear atlas building approach.
November 7, 2005 T-Con notes (R Gollub) In Attendence (and all on time!) S. Pieper, S. Mori, J. Fallon, M. Perry, R. Gollub, M. Panzenboeck, A. Caprihan (Univ New Mexico), Vid Somebody (UCI)
Progress: All 3 sites have received a DVD from Susumu with six data sets (subjects: 2, 3, and 5 and another subject who was scanned three times 4_1, 4_2, 4_3) and have begun to practice doing the tractography following the methods in his manuscript draft. Agreed upon these additional points:
- By the end of this week each of the three data analysis folks, Michelle, Martina, Arvind and Jim (possibly Vid at UCI will also participate as a 4th separate person) will load data from subject 4_1 and follow the instructions to isolate the first track, the cingulum bundle. They will save the tracking result in a binary format (256x256x50 matrix filled with zeros except pixels with the identified fibers which are ones, file size ~4MB) and send it to Susumu for him to verify that they understand the directions and that he can read the files they generate.
- By December 1st each of the analysis folks will complete 2 full sets of DTI fiber tracks (11 separate tracks) on subject 4_1 and send them to Susumu. The first set will use the method of specifying a first ROI with an "or" command and a second ROI with an "and" command to isolate the track. The second set will use the method of specifying the "cut" command that waits for the two ROIs to be specified. Susumu is interested to compare which method works better. To be fair, the three or four of you should randomly divide yourselves into two groups differing on which of the two methods you do first so that the results of this mini comparison are not confounded by experience.
- We will have the next T-con on Friday Dec. 9th at 2 PM EST to review the results and to give each of the analysis folks feedback.
- At that next call we will set the deadline (hopefully before January 1st) for each of the data analysis folks to complete subjects 2, 3 and 5.
No information on the progress of the generation of NRRD readers for Susumu's data. Steve Pieper to follow up with Jonathan Farrell on this. Target deadline is December 9th to have this completed for all the DTI datasets that Susumu has posted on his own website.
October 17-18, 2005 Discussions at the All Hands Meeting (S. Pieper, S. Mori, A. Dale, J. Fallon, E. Halgren, C. Fennema-Notestine, M. Perry, K. Lim, A. Song, R. Gollub, M. Panzenboeck, J. MacFall, S. Czanner, J. Jovicich, W. Kremen, J. Farrell, and Bin Chen)
See also DTI Calibration Core pages for updates on the progress and results from MRI acquisition parameter exploration.
The group enthusiastically embraced the concept of performing a focused set of validation experiments on DTI image analysis and proposed that this effort be extended and supported by our collaborative efforts with NA-MIC.
The sequence of studies proposed is:
'Project Number 1A: "Assessment of the reliability of an established tractography method"'
Goals:
- Determine the reproducibility of DTI tractography measures across different investigators using the same preprocessed data and the same tractography tool and specifications for placement of manual ROIs
- Allow our BIRN neuroanatomist to verify the ROI definitions and determine if any additional pathways need to be identified for the purposes of our clinical collaborators (e.g. VETSA and MIND studies)
- Train BIRN and collaborative investigators in the use of DTI Studio tractography tool
- Provide feedback to S. Mori regarding helpful improvements in the text of his manual
Timeline:
- Immediate establish an email list for this working group and set up a teleconference call to coordinate the work (Randy)
- October 31, 2005 Susumu will get "David data" on CD to Steve Pieper for distribution to our working group
- November 21, 2005 all three investigators complete the tractography on "David data" so inter rater reliability data can be included in his manuscript
- January 1, 2006 all three investigators complete the tractography measures on the first 10 shared data sets
- January 9-10, 2006 development of statistical analysis methods collaboratively with NA-MIC at their AHM
- March 2006 Completion of study and manuscript preparation (Susumu already has this well underway)
Methods:
- Three investigators have been confirmed, each of whom is also the key point person for the analysis of other large collaborating clinical research studies indicated in brackets: Michelle Perry (VETSA), Martina Panzenboeck (NA-MIC Core 3), Arvind Caprihan (MIND)
- Each of them will download the DTI Studio Software and the first 10 public DTI processed data sets (FA data (*.ani) and VECTOR data (*.vec) from the JHU Lab of Brain Anatomical MRI webpage [1] and download the ROI descriptions and Figures from the SPL sponsored M-BIRN xythos files sharing site (https://share.spl.harvard.edu/xythoswfs/webui) in the folder named DTI Analysis Calibration.
- These three investigators will replicate the tractography measures on the David and n=10 public data sets following S. Mori's exact methods as described in the UserManual_V2_10_6.doc from the website and the draft manuscript.
- Outcome measures to be collected by each of the three investigators include:
1. Expert neuroanatomist visually inspects and certifies the results Question: (methods are vague- to be clarified) this will most likely be J. Fallon unless the NA-MIC Core 3 folks decide to participate 2. NA-MIC tool fiber tract similarity metric (bundle differences), tract reproducibility metric
Project Number 1B:
Identify additional tracts (to be determined during a working session at the M-BIRN AHM in Sedona in March) Validate them
Project Number 2: Validation of DTI methods on the MIND Reliability data (n = 10, 4 sites, test-retest at each site, 80 data sets) to be done in collaboration with NA-MIC Core 1 investigators
Additional work to be articulated at the NA-MIC AHM in January 2006 (tenative plan to be verified in near future)
August 4, 2005 (S. Pieper)
- A phone conference including Anders Dale, Steve Pieper, Steve Potkin, Jim Fallon, Martina Panzen was held to review status of the white matter atlas generation at UCI and its application to the VETSA project.
- It was agreed that there are two types of atlas work:
- a label map based white matter atlas of the type being generated by Susumu Mori at JHU, which it is hoped will be applicable to data through direct registration
- a tractography based atlas which will be a longer term project
- UCI expressed concern that the Slicer-based tractography atlas they had previously generated would be difficult to reproduce across multiple subjects. The group agreed to investigate the issues and confirm that newer versions of the software behave the same as earlier releases or if new issues have caused scaling problems. UCI and BWH will investigate this. UCI will also review their techniques and explore various tools to ensure that they can deliver their white matter atlas committment.
July 27, 2005 (S. Pieper)
- A meeting was held in San Diego with Dave Kennedy, Steve Pieper meeting with Anders Dale's research group. We reviewed the white matter atlas from Susumu Mori and the current test datasets from VETSA. It looks like all the pieces are in place to try doing registration and Steve Pieper is testing the tensor-based registration code in the current version of slicer.
- Duke has identified test subjects for the BELL tests on MIRIAD data. These non-tensor registrations will also be tested by Steve Pieper before the October meeting.
May 17, 2005 (J. Jovicich)
- Probabilistic DTI-based White Matter Label Atlas
- Susumu is building a white matter probabilistic atlas for use in the MIRIAD data. This collaboration was initiated in March-Miami meeting by Steve and Jim.
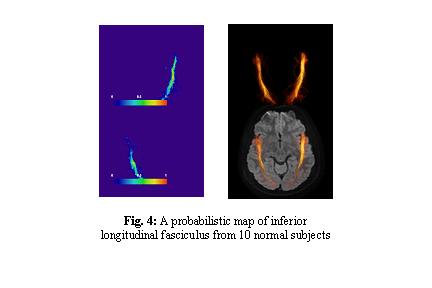
- The idea is to create probabilistic maps of individual white matter tracts using tractography and to use the coordinate information to answer the following hypothesis; "Can we superimpose the coordinate information on existing MRI to correlate MRI findings (e.g. T2 lesions, fMRI activation maps) with specific white matter tracts?"
- We finished creation of such probabilistic maps for various white matter tracts (Fig. 4) using data from 10 healthy volunteers. Currently, they are in JHU DTI template coordinates.
- Pending issues are;
- We'll transform the data to ICBM-MNI coordinates to adopt more widely-used coordinates
- We are increasing the number from 10 to 30 volunteers.
- We'll finish these two issues in May and send them to Jorge.
- By mid-June the atlas will consist of 30 averaged subjects and the coordinates will be in ICBM-MRI. This will then be passed on to Steve Pieper for co-registration with the MIRIAD data and then to James MacFall for the lesion/tractography analysis.
- Probabilistic DTI-based Tractography Atlas
- Building a tractography atlas from the VETSA data: J. Fallon has the 3D Slicer tools ready to work on the VETSA dataset. NEED: i) Anders to provide VETSA data with structural data co-registered with diffusion data, in nrrd format; ii) white matter atlas from Dave Kennedy to help map tracks that are closer to cortex.
February 14, 2005 (J. Jovicich)
- Steve Pieper needs to characterize diffusion MRI format variability across mBIRN & fBIRN so that he can extend 3D Slicer to allow flexible data importing and DTI visualization. Jorge sent out request for mBIRN and fBIRN sites (see also Acquisition/analysis of test-retest DTI, Updates from Feb 14, 2005).
- This week Steve and Raul Estepar will be at UCI assisting James & Michelle with their use of 3D Slicer to load DTI data and build track atlas efficiently.
February 3, 2005 (S. Pieper)
- Continued close interaction between J. Fallon, M. Perry, S. Pieper, and R. Estepar on using the DTMRI module in slicer for creating white matter atlases from scans obtained using the MGH protocol.