Difference between revisions of "Projects:RegistrationDocumentation:ParameterTesting"
From NAMIC Wiki
| Line 18: | Line 18: | ||
**range of fiducial misalignment & distributions | **range of fiducial misalignment & distributions | ||
**Plot error vs. initial misalignment (where does registration begin to fail), plot error vs. parameter settings (which setting works best for the toughest case) | **Plot error vs. initial misalignment (where does registration begin to fail), plot error vs. parameter settings (which setting works best for the toughest case) | ||
| + | **Examples below are cost function comparisons: '''MI''' vs '''NCorr''' | ||
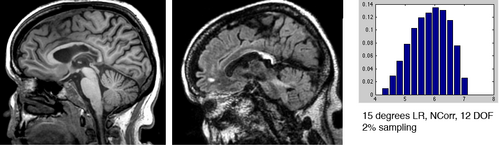
[[Image:SRegTest_FLAIR-T1_LRRot15.png|left|500px|360 Level Self Validation Test: FLAIR to T1, left-right rotation]] | [[Image:SRegTest_FLAIR-T1_LRRot15.png|left|500px|360 Level Self Validation Test: FLAIR to T1, left-right rotation]] | ||
[[Image:RegistrationISMRM10_Disturb_12_hist.png|left|300px|RMS Histogram comparison for 12 degrees misalignment]] | [[Image:RegistrationISMRM10_Disturb_12_hist.png|left|300px|RMS Histogram comparison for 12 degrees misalignment]] | ||
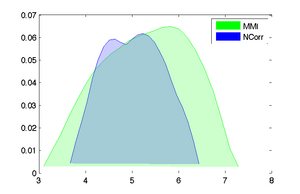
[[Image:RegistrationISMRM10_RMS.png|left|300px|Sensitivity (RMS range) vs. rotational misalignment]] | [[Image:RegistrationISMRM10_RMS.png|left|300px|Sensitivity (RMS range) vs. rotational misalignment]] | ||
Revision as of 22:11, 27 October 2009
Home < Projects:RegistrationDocumentation:ParameterTestingBack to ARRA main page
Back to Registration main page
Back to Registration Use-case Inventory
ISMRM abstract 2010
- Title: MR-protocol Tailored Medical Image Registration
- Objective: Determine optimized sets of parameters for successful automated registration of MR-MR images within the 3DSlicer software. DOF, cost function, initialization and optimization strategy will differ because of the differences in image contrast and/or content. This work will present approaches and solutions for successful registration for a large set of combinations of MRI pairings. This is part of a concerted effort to build a Registration Case Library available to the medical imaging research community.
- Method:
- 1- choose 3-4 subjects/exams with 3-4 different contrast pairings: T1, T2, PD, FLAIR: ~12-16 images. We choose sets for which we have a good registration solution
- 2- disturb each pair by a known transform of varying rotational & translational misalignment
- 3- run registration for a set of parameter settings and save the result Xform, e.g. metric: NormCorr vs. MI , 2% vs 5% sampling, 50 vs. 100 iteration max
- 4- evaluate registration error as point distance and RMS.
- 5- run sensitivity analysis and report the best performing parameter set for each MR-MR combination
- 6-extension 1: add different voxel sizes, i.e. emulate 1,3,5mm slice thickness
- This self-validation scheme avoids recruiting an expert reader to determine ~ 3-5 anatomical landmarks on each unregistered image pair (time constraint). Also we can cover a wider range of misalignment and sensitivity by controlling the input Xform. It also facilitates batch processing.
- Results:
- range of fiducial misalignment & distributions
- Plot error vs. initial misalignment (where does registration begin to fail), plot error vs. parameter settings (which setting works best for the toughest case)
- Examples below are cost function comparisons: MI vs NCorr