Difference between revisions of "2009 Summer Project Week Slicer3 Brainlab Introduction"
From NAMIC Wiki
(Created page with '__NOTOC__ <gallery> Image:PW2009-v3.png|Project Week Main Page Image:genuFAp.jpg|Scatter plot of the original FA data through the genu of the corpus ...') |
m (Text replacement - "http://www.slicer.org/slicerWiki/index.php/" to "https://www.slicer.org/wiki/") |
||
| (17 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
<gallery> | <gallery> | ||
| − | Image:PW2009-v3.png|[[2009_Summer_Project_Week|Project Week Main Page]] | + | Image:PW2009-v3.png|[[2009_Summer_Project_Week#Projects|Project Week Main Page]] |
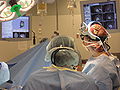
| − | Image: | + | Image:Slicer_dti_seeding_or.jpg|Slicer3 in OR for fiducial seeding of DTI tractography |
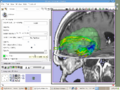
| − | Image: | + | Image:Slicer_dti_seeding.png|Slicer3 fiducial seeding of DTI tractography |
</gallery> | </gallery> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
==Key Investigators== | ==Key Investigators== | ||
| − | * | + | * BWH: Haiying Liu, Isaiah Norton, Junichi Tokuda, Alex Golby, Noby Hata, Tina Kapur, Haytham Elhawary |
| − | * | + | * Isomics: Steve Pieper |
| − | + | * Yale: Xenios Papademetris | |
| + | * BrainLab: Pratik Patel | ||
| + | * UCLA: Nathan Hageman | ||
<div style="margin: 20px;"> | <div style="margin: 20px;"> | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
| − | + | Introduce and demonstrate how we integrate BrainLab, BioImage Suite and Slicer3 to perform fiducial seeding of DTI tractography in Slicer3. | |
</div> | </div> | ||
| Line 28: | Line 23: | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| − | + | *Slicer3, BioImage Suite and BrainLab will be connected through a router in BWH OR. BioImage Suite will be a "bridge" between Slicer3 and BrainLab for data communication. | |
| − | + | *It's probably a good idea to show step by step how we set up the system. Instructions can also be found here at: https://www.slicer.org/wiki/Slicer3:BrainLab_Integration | |
| − | |||
</div> | </div> | ||
| Line 36: | Line 30: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | + | * Performed demonstration (with great help from Yale team and BrainLab) this week. | |
| − | + | * Five participants. | |
| + | * Launching BWH-UCLA collaboration based this work. | ||
</div> | </div> | ||
</div> | </div> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Latest revision as of 17:23, 10 July 2017
Home < 2009 Summer Project Week Slicer3 Brainlab IntroductionKey Investigators
- BWH: Haiying Liu, Isaiah Norton, Junichi Tokuda, Alex Golby, Noby Hata, Tina Kapur, Haytham Elhawary
- Isomics: Steve Pieper
- Yale: Xenios Papademetris
- BrainLab: Pratik Patel
- UCLA: Nathan Hageman
Objective
Introduce and demonstrate how we integrate BrainLab, BioImage Suite and Slicer3 to perform fiducial seeding of DTI tractography in Slicer3.
Approach, Plan
- Slicer3, BioImage Suite and BrainLab will be connected through a router in BWH OR. BioImage Suite will be a "bridge" between Slicer3 and BrainLab for data communication.
- It's probably a good idea to show step by step how we set up the system. Instructions can also be found here at: https://www.slicer.org/wiki/Slicer3:BrainLab_Integration
Progress
- Performed demonstration (with great help from Yale team and BrainLab) this week.
- Five participants.
- Launching BWH-UCLA collaboration based this work.