Difference between revisions of "DBP2:HarvardFinal:2010"
| (9 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
= Short overview = | = Short overview = | ||
| − | The main goal of this project is to develop end-to-end application that would be used to characterize anatomical connectivity abnormalities in the brain of patients with velocardiofacial syndrome (VCFS), and to link this information with deficits in schizophrenia. | + | The main goal of this project is to develop end-to-end application that would be used to characterize anatomical connectivity abnormalities in the brain of patients with velocardiofacial syndrome (VCFS), and to link this information with deficits in schizophrenia. During the duration of this project, we developed, disseminated and applied to clinical studies software containing method based on stochastic tractography. |
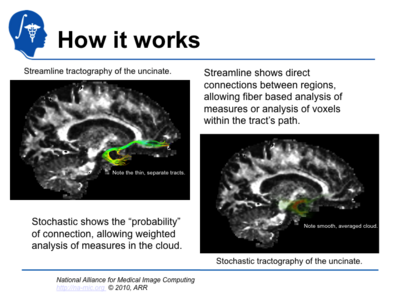
| + | Stochastic Tractography has been developed to quantify the uncertainty associated with estimated fiber tracts (Bjornemo et al., 2002). Method uses a propagation model based on stochastics and regularization, which allows paths originating at one point to branch and return a probability distribution of possible paths. The method utilizes principles of a statistical Monte Carlo method called Sequential Importance Sampling and Resampling (SISR). Based on probability functions, using a sequential importance sampling technique (Bjornemo et al., 2002), one can generate thousands of fibers starting in the same point by sequentially drawing random step directions. This gives a very rich model of the fiber distribution, as contrasted with single fibers produced by conventional tractography methods. Moreover, from a large number of sampled paths, probability maps can be generated, providing better estimates of connectivity between several anatomical locations. | ||
| + | [[Image:Slide08.png|thumb|right|400px|]] | ||
= Related publications = | = Related publications = | ||
| Line 27: | Line 29: | ||
= Listing and short description of the software that will run in Slicer 3.6.1 and what it will accomplish = | = Listing and short description of the software that will run in Slicer 3.6.1 and what it will accomplish = | ||
| + | |||
| + | Stochastic Tractography module is currently part of Slicer 3.6 release. The software, when provided two "seeding" regions, finds the probability of connection between those regions, by calculating streamline tracts with a small amount of variation introduced at each step. These tracts are then averaged, to produce a "stochastic" cloud. The module is currently python based. Module is flexible, containing options for data smoothing, filtering, thresholding, changing tractography settings (such as total number of tracts, maximum length, step size, spacing, stopping), as well as types of output (connectivity map, FA, Mode, Trace). Inputs and outputs are compatible with other slicer modules, and can be visualized in Slicer (including 3D volume rendering). End to end tutorial for running stochastic tractography can be found here: | ||
| + | |||
| + | Tutorial ([[Media:Stochastic Tractography TutorialContestSummer2010.pdf|PDF - 1MB]]) | ||
| + | |||
| + | Tutorial ([[Media:Stochastic_Tractography_TutorialContestSummer2010.ppt|PPT - 1.2MB]]) | ||
| + | |||
= Listing and short description of the sample data = | = Listing and short description of the sample data = | ||
| + | Structural MRI (sMRI). For the Structural MRI volume measures, images were acquired using a 3T GE scanner at BWH in Boston, MA. We used an 8 Channel coil and ASSET with a SENSE-factor of 2. The structural MRI acquisition protocol includes contiguous spoiled gradient-recalled acquisition (fastSPGR) with the following parameters; TR=7.4ms, TE=3ms, TI=600, 10 degree flip angle, 25.6cm2 field of view, matrix=256x256. The voxel dimensions are 1x1x1 mm. | ||
| + | Diffusion Tensor Imaging (DTI). We used an echo planar imaging (EPI) DTI Tensor sequence. We used a double echo option to reduce eddy-current related distortions (Heid 2000; Alexander 1997). To reduce impact of EPI spatial distortion, we used an 8 Channel coil and ASSET with a SENSE-factor of 2. We acquired 51 directions with b=900, 8 baseline scans with b=0. Scan parameters are: TR 17000 ms, TE 78 ms, FOV 24 cm, 144x144 encoding steps, 1.7 mm slice thickness. 85 axial slices parallel to the AC-PC line cover the whole brain. | ||
| + | Sample data set can be found here: | ||
| + | |||
| + | Dataset ([[Media:Stochastic_tutorial_data_TutorialContestSummer2010.zip|ZIP - 92MB]]) | ||
Latest revision as of 18:04, 12 November 2010
Home < DBP2:HarvardFinal:2010back to DBP2 Main
Contents
Short overview
The main goal of this project is to develop end-to-end application that would be used to characterize anatomical connectivity abnormalities in the brain of patients with velocardiofacial syndrome (VCFS), and to link this information with deficits in schizophrenia. During the duration of this project, we developed, disseminated and applied to clinical studies software containing method based on stochastic tractography. Stochastic Tractography has been developed to quantify the uncertainty associated with estimated fiber tracts (Bjornemo et al., 2002). Method uses a propagation model based on stochastics and regularization, which allows paths originating at one point to branch and return a probability distribution of possible paths. The method utilizes principles of a statistical Monte Carlo method called Sequential Importance Sampling and Resampling (SISR). Based on probability functions, using a sequential importance sampling technique (Bjornemo et al., 2002), one can generate thousands of fibers starting in the same point by sequentially drawing random step directions. This gives a very rich model of the fiber distribution, as contrasted with single fibers produced by conventional tractography methods. Moreover, from a large number of sampled paths, probability maps can be generated, providing better estimates of connectivity between several anatomical locations.
Related publications
- Björnemo M, Brun A, Kikinis R, Westin CF. Regularized stochastic white matter tractography using diffusion tensor MRI. In Fifth International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI'02). Tokyo, Japan, 2002;435-442.
- Friman, O., Farneback, G., Westin CF. A Bayesian Approach for Stochastic White Matter Tractography. IEEE Transactions on Medical Imaging, Vol 25, No. 8, Aug. 2006
- Melonakos J, Mohan V, Niethammer M, Smith K, Kubicki M, Tannenbaum A. (2007): Finsler tractography for white matter connectivity analysis of the cingulum bundle. Med Image Comput Comput Assist Interv Int Conf Med Image Comput Comput Assist Interv 10(Pt 1):36-43.
- Rosenberger G, Kubicki M, Nestor PG, Connor E, Bushell GB, Markant D, Niznikiewicz M, Westin CF, Kikinis R, J Saykin A, McCarley RW, Shenton ME. (2008): Age-related deficits in fronto-temporal connections in schizophrenia: a diffusion tensor imaging study. Schizophr Res Jul;102(1-3):181-188.
- Nestor PG, Kubicki M, Niznikiewicz M, Gurrera RJ, McCarley RW, Shenton ME. (2008): Neuropsychological disturbance in schizophrenia: a diffusion tensor imaging study. Neuropsychology Mar;22(2):246-254.
- Aja-Fernandez S, Niethammer M, Kubicki M, Shenton, ME, Westin, C-F. (2008): Restoration of DWI Data Using a Rician LMMSE Estimator. IEEE Trans Med Imaging Oct; 27(10):1389-403.
- Kubicki M, Styner M, Bouix S, Gerig G, Markant D, Smith K, Kikinis R, McCarley RW, Shenton ME. (2008): Reduced Interhemispheric Connectivity in Schizophrenia- Tractography Based Segmentation of the Corpus Callosum. Schizophr Res 106(2 3):125-31.
- Maddah M, Kubicki M, Wells WM, Westin CF, Shenton ME, Grimson WE. (2008): Findings in schizophrenia by tract-oriented DT-MRI analysis. Med Image Comput Comput Assist Interv Int Conf Med Image Comput Comput Assist Interv (MICCAI) 11(Pt 1):917-24.
- Fitzsimmons J, Kubicki M, Smith K, Bushell G, Estepar RS, Westin CF, Nestor PG, Niznikiewicz MA, Kikinis R, McCarley RW, Shenton ME. (2009): Diffusion tractography of the fornix in schizophrenia. Schizophr Res Jan;107(1):39-46.
- Lee K, Yoshida T, Kubicki M, Bouix S, Westin CF, Kindlmann G, Niznikiewicz M, Cohen A, McCarley RW, Shenton ME. (2009): Increased diffusivity in superior temporal gyrus in patients with schizophrenia: A Diffusion Tensor Imaging study. Schizophr Res Jan 8. [Epub ahead of print].
- Kawashima T, Nakamura M, Bouix S, Kubicki M, Salisbury D, Westin CF, McCarley RW, Shenton ME. (2009): Uncinate fasciculus abnormalities in recent onset schizophrenia and affective psychosis: A diffusion tensor imaging study: Schizophr Res 110: 119-126
- Oh JS, Kubicki M, Rosenberger G, Bouix S, Levitt JL, McCarley RW, Westin C-F, Shenton ME. (2009): Thalamo-Frontal White Matter Alterations in Chronic Schizophrenia: A Quantitative Diffusion Tractography Study. Hum Brain Mapp. Nov;30(11):3812-25.
- Jeong BS, Wible CG, Hashimoto RH, Kubicki M. (2009): Functional and Anatomical Connectivity Abnormalities in Left Inferior Frontal Gyrus in Schizophrenia. Hum Brain Mapp. Dec;30(12):4138-51.
- Ungar L, Niznikiewicz M, Nestor P, Kubicki M. (2010): Color Stroop and Negative Priming in Schizophrenia: An fMRI Study. Psychiatry Res Jan 30;181(1):24-9.
- Jeong BS, Kubicki M. (2010): Reduced Task-related Suppression during Semantic Repetition Priming in Schizophrenia. Psychiatry Res. Feb 28;181(2):114-20.
- Nestor PG, Kubicki M, Nakamura M, Niznikiewicz M, McCarley RW, Shenton ME. (2010): Comparing prefrontal gray and white matter contributions to intelligence and decision making in schizophrenia and healthy controls. Neuropsychology. Jan;24(1):121-9.
- Whitford TJ, Kubicki M, Schneiderman JS, O'Donnell LJ, King R, Alvarado JL, Khan U, Markant D, Nestor PG, Niznikiewicz M, McCarley RW, Westin CF, Shenton ME. (2010): Corpus Callosum Abnormalities and Their Association with Psychotic Symptoms in Patients with Schizophrenia. Biol Psychiatry. May 20. [Epub ahead of print]
- Kubicki M, Niznikiewicz M, Connor E, Ungar L, Nestor PG, Bouix S, Dreusicke M, Kikinis R, McCarley RW, Shenton ME. (In Press): Relationship Between White Matter Integrity, Attention, and Memory in Schizophrenia: A Diffusion Tensor Imaging Study. Brain Imaging and Behavior.
Listing and short description of the software that will run in Slicer 3.6.1 and what it will accomplish
Stochastic Tractography module is currently part of Slicer 3.6 release. The software, when provided two "seeding" regions, finds the probability of connection between those regions, by calculating streamline tracts with a small amount of variation introduced at each step. These tracts are then averaged, to produce a "stochastic" cloud. The module is currently python based. Module is flexible, containing options for data smoothing, filtering, thresholding, changing tractography settings (such as total number of tracts, maximum length, step size, spacing, stopping), as well as types of output (connectivity map, FA, Mode, Trace). Inputs and outputs are compatible with other slicer modules, and can be visualized in Slicer (including 3D volume rendering). End to end tutorial for running stochastic tractography can be found here:
Tutorial (PDF - 1MB)
Tutorial (PPT - 1.2MB)
Listing and short description of the sample data
Structural MRI (sMRI). For the Structural MRI volume measures, images were acquired using a 3T GE scanner at BWH in Boston, MA. We used an 8 Channel coil and ASSET with a SENSE-factor of 2. The structural MRI acquisition protocol includes contiguous spoiled gradient-recalled acquisition (fastSPGR) with the following parameters; TR=7.4ms, TE=3ms, TI=600, 10 degree flip angle, 25.6cm2 field of view, matrix=256x256. The voxel dimensions are 1x1x1 mm. Diffusion Tensor Imaging (DTI). We used an echo planar imaging (EPI) DTI Tensor sequence. We used a double echo option to reduce eddy-current related distortions (Heid 2000; Alexander 1997). To reduce impact of EPI spatial distortion, we used an 8 Channel coil and ASSET with a SENSE-factor of 2. We acquired 51 directions with b=900, 8 baseline scans with b=0. Scan parameters are: TR 17000 ms, TE 78 ms, FOV 24 cm, 144x144 encoding steps, 1.7 mm slice thickness. 85 axial slices parallel to the AC-PC line cover the whole brain. Sample data set can be found here:
Dataset (ZIP - 92MB)