Difference between revisions of "2014 Project Week:GraphCutsLASegmentationModule"
From NAMIC Wiki
(Added progress) |
|||
| (4 intermediate revisions by one other user not shown) | |||
| Line 6: | Line 6: | ||
Josh Cates (CARMA center, University of Utah) | Josh Cates (CARMA center, University of Utah) | ||
| + | |||
| + | Rob Macleod (CARMA center, SCI Institute, University of Utah) | ||
Ross Whitaker(SCI Institute, University of Utah) | Ross Whitaker(SCI Institute, University of Utah) | ||
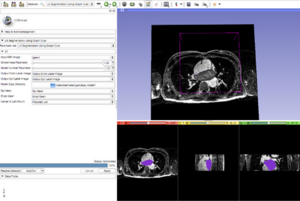
| − | + | [[File:LAsegment GraphCut.png|thumb|center|300px]] | |
==Project Description== | ==Project Description== | ||
| Line 14: | Line 16: | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
| − | * | + | * Develop a Slicer module that automatically segments the left atrial wall from a given LGE-MRI image. |
</div> | </div> | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| − | * | + | * Involves Bayesian formulation with Markov random field prior within a nested-layer 3D mesh which leads to surface-net problem [Veni ''et al'', IPMI 2013]. |
| + | * Solved by using VCEnet strategy and graph-cuts [Wu and Chen, 2002]. | ||
| + | * Uses training strategy in order to generate model shapes as well as to compute costs at each mesh point. | ||
</div> | </div> | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | * | + | * The module now provides the ability to the user to select the approximate center of the left atrium and the model is aligned with that point. |
| + | * The VCEnet technique produces better results than the Vnet technique that was used in previous versions of the module. | ||
| + | * Module is now available in the Slicer. It could be downloaded from: https://github.com/carma-center/carma_slicer_extension/ | ||
| + | * The online documentation on its usage is available at: https://www.slicer.org/slicerWiki/index.php/Documentation/Nightly/Modules/AutomatedLASegmentation | ||
| + | * Model data could be downloaded from: http://slicer.kitware.com/midas3/folder/1550 | ||
| + | |||
</div> | </div> | ||
</div> | </div> | ||
Latest revision as of 03:04, 10 January 2014
Home < 2014 Project Week:GraphCutsLASegmentationModuleKey Investigators
Gopalkrishna Veni (SCI Institute, University of Utah)
Salma Bengali (CARMA center, University of Utah)
Josh Cates (CARMA center, University of Utah)
Rob Macleod (CARMA center, SCI Institute, University of Utah)
Ross Whitaker(SCI Institute, University of Utah)
Project Description
Objective
- Develop a Slicer module that automatically segments the left atrial wall from a given LGE-MRI image.
Approach, Plan
- Involves Bayesian formulation with Markov random field prior within a nested-layer 3D mesh which leads to surface-net problem [Veni et al, IPMI 2013].
- Solved by using VCEnet strategy and graph-cuts [Wu and Chen, 2002].
- Uses training strategy in order to generate model shapes as well as to compute costs at each mesh point.
Progress
- The module now provides the ability to the user to select the approximate center of the left atrium and the model is aligned with that point.
- The VCEnet technique produces better results than the Vnet technique that was used in previous versions of the module.
- Module is now available in the Slicer. It could be downloaded from: https://github.com/carma-center/carma_slicer_extension/
- The online documentation on its usage is available at: https://www.slicer.org/slicerWiki/index.php/Documentation/Nightly/Modules/AutomatedLASegmentation
- Model data could be downloaded from: http://slicer.kitware.com/midas3/folder/1550