Difference between revisions of "DBP2:Harvard:Brain Segmentation Roadmap"
| Line 8: | Line 8: | ||
== Algorythm == | == Algorythm == | ||
| − | {|cellpadding=" | + | {|cellpadding="0" |
| style="width:85%" | | | style="width:85%" | | ||
| − | + | ; A-Description | |
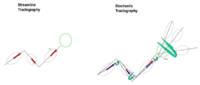
| − | + | * Most tractography methods estimate fibers by tracing the maximum direction of diffusion. A limitation of this approach is that, in practice, several factors introduce uncertainty in the tracking procedure, including, noise, splitting and crossing fibers, head motion and image artifacts. To address this uncertainty, stochastic tractography methods have been developed to quantify the uncertainty associated with estimated fibers (Bjornemo et al., 2002). Method uses a propagation model based on stochastics and regularization, which allows paths originating at one point to branch and return a probability distribution of possible paths. The method utilizes principles of a statistical Monte Carlo method called Sequential Importance Sampling and Resampling (SISR). Based on probability functions, using a sequential importance sampling technique ([http://lmi.bwh.harvard.edu/papers/pdfs/2002/bjornemoMICCAI02.pdf Bjornemo et al., 2002]), one can generate thousands of fibers starting in the same point by sequentially drawing random step directions. This gives a very rich model of the fiber distribution, as contrasted with single fibers produced by conventional tractography methods. Moreover, from a large number of sampled paths, probability maps can be generated, providing better estimates of connectivity between several anatomical locations. A comparison of the algorithms can be see here. (Figure 1) | |
| style="width:15%" | [[Image:IC_sto_new.png|thumb|right|200px|<font size=1> Figure 1: Comparison of deterministic and stochastic tractography algorithms</font>]] | | style="width:15%" | [[Image:IC_sto_new.png|thumb|right|200px|<font size=1> Figure 1: Comparison of deterministic and stochastic tractography algorithms</font>]] | ||
|} | |} | ||
| − | {|cellpadding=" | + | {|cellpadding="0" |
| style="width:85%" | | | style="width:85%" | | ||
| − | + | ; B-Possible Applications | |
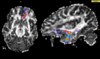
| − | + | * Since diffusion direction uncertainty within the gray matter is quite significant; principal diffusion direction approaches usually do not work for tracking between two gray matter regions. Thus if one requires finding connections between a priori selected anatomical gray matter regions, defined either by anatomical segmentations (in case of using structural ROI data), or functional activations (in case of megring DTI with fMRI), stochastic tractography seems to be the method of choice. Here is an example of this application to anatomical data (Figure 2) and to fMRI data (Figure 3). | |
| − | + | * Stochastic Tractography is also comparable, if not better, in defining large white matter fiber bundles, especially those traveling through white matter regions characterized by increased diffusion uncertainty (fiber crossings). Example of such application to internal capsule. (Figure 4) | |
| − | | style="width: | + | | style="width:7.5%" | [[Image:OFC.png|thumb|right|100px|<font size=1>Figure 2: Connection between OFC and amygdala</font>]] |
| + | | style="width:7.5%" | [[Image:Cingulate_gyrus.jpg|thumb|right|100px|<font size=1>Figure 3: Connection between anterior and posterior cingulate gyrus fMRI activations</font>]] | ||
|} | |} | ||
| − | + | [[Image:IC-comp-new.png|thumb|right|200px|<font size=1>Figure 4: Streamline vs. Stochastic Tractography of the Internal Capsule</font>]] | |
| − | + | ; C-References | |
| − | |||
| − | == Module == | + | * [http://lmi.bwh.harvard.edu/papers/pdfs/2002/bjornemoMICCAI02.pdf Björnemo M, Brun A, Kikinis R, Westin CF. Regularized stochastic white matter tractography using diffusion tensor MRI. In Fifth International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI'02). Tokyo, Japan, 2002;435-442.] |
| + | * [http://lmi.bwh.harvard.edu/papers/pdfs/2006/frimanTMI06.pdf Friman, O., Farneback, G., Westin CF. A Bayesian Approach for Stochastic White Matter Tractography. IEEE Transactions on Medical Imaging, Vol 25, No. 8, Aug. 2006] | ||
| + | |||
| + | ==Module== | ||
Fill it in once its ready. Write exactly, step by step what is its functionality, adding snapshots: | Fill it in once its ready. Write exactly, step by step what is its functionality, adding snapshots: | ||
| Line 57: | Line 60: | ||
:**[[Media:SlicerSTtutorial.ogg|Training Webcast]] ([http://www.theora.org Theora format]) | :**[[Media:SlicerSTtutorial.ogg|Training Webcast]] ([http://www.theora.org Theora format]) | ||
| − | == Work Accomplished == | + | ==Work Accomplished== |
| − | + | {|cellpadding="0" | |
| + | | style="width:85%" | | ||
; A - Optimization and testing of stochastic tractography algorythm : | ; A - Optimization and testing of stochastic tractography algorythm : | ||
| − | |||
:* Original methodological paper, as well as our first attempts to use the algorythm (CC+ and matlab scripts) have been done on old "NAMIC" 1.5T LSDI data ([https://portal.nbirn.net:443/gridsphere/gridsphere?cid=srbfilebrowser&gs_action=gotoDirectory&CPq_dirpath=%2Fhome%2FProjects%2FNAMIC__0003%2FFiles%2FHarvard%2Farchive_morph&up=CPq&JavaScript=enabled Structural MRI] and [https://portal.nbirn.net:443/gridsphere/gridsphere?cid=srbfilebrowser&gs_action=gotoDirectory&up=7li&7li_dirpath=%2Fhome%2FProjects%2FNAMIC__0003%2FFiles%2FHarvard%2Farchive_diffusion&JavaScript=enabled DTI data]). | :* Original methodological paper, as well as our first attempts to use the algorythm (CC+ and matlab scripts) have been done on old "NAMIC" 1.5T LSDI data ([https://portal.nbirn.net:443/gridsphere/gridsphere?cid=srbfilebrowser&gs_action=gotoDirectory&CPq_dirpath=%2Fhome%2FProjects%2FNAMIC__0003%2FFiles%2FHarvard%2Farchive_morph&up=CPq&JavaScript=enabled Structural MRI] and [https://portal.nbirn.net:443/gridsphere/gridsphere?cid=srbfilebrowser&gs_action=gotoDirectory&up=7li&7li_dirpath=%2Fhome%2FProjects%2FNAMIC__0003%2FFiles%2FHarvard%2Farchive_diffusion&JavaScript=enabled DTI data]). | ||
:* Tri worked hard on making sure algorythm works on new high resolution 3T data (available here: [https://portal.nbirn.net/gridsphere/gridsphere?cid=srbfilebrowser&gs_action=moveUpDir&gv7_dirpath=%2Fhome%2FProjects%2FNAMIC__0003%2FFiles%2FPNL%2F3T_strct_dti_fmri%2Fcase01017&up=gv7&JavaScript=enabled|PNL 3T Data]). | :* Tri worked hard on making sure algorythm works on new high resolution 3T data (available here: [https://portal.nbirn.net/gridsphere/gridsphere?cid=srbfilebrowser&gs_action=moveUpDir&gv7_dirpath=%2Fhome%2FProjects%2FNAMIC__0003%2FFiles%2FPNL%2F3T_strct_dti_fmri%2Fcase01017&up=gv7&JavaScript=enabled|PNL 3T Data]). | ||
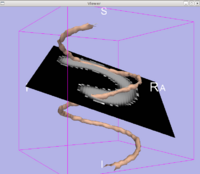
:* Tests have been done also on the spiral diffusion phantom, to make sure diffusion directions and scanner coordinates are handled properly by the algorythm (Figure 5). | :* Tests have been done also on the spiral diffusion phantom, to make sure diffusion directions and scanner coordinates are handled properly by the algorythm (Figure 5). | ||
| − | + | | style="width:15%" | [[Image:spiral2.png|thumb|right|200px|<font size=1>Figure 5: Stochastic Tractography on Spiral Phantom</font>]] | |
| + | |} | ||
| + | {|cellpadding="0" | ||
| + | | style="width:85%" | | ||
; B - Clincal Applications | ; B - Clincal Applications | ||
| Line 82: | Line 88: | ||
; B - Related Clinical Projects | ; B - Related Clinical Projects | ||
| − | + | ||
| + | | style="width:85%" | | ||
| + | |||
| + | |} | ||
| + | |||
| + | {|cellpadding="0" | ||
| + | |||
| + | | style="width:85%" | | ||
;* Arcuate Fasciculus Extraction Project | ;* Arcuate Fasciculus Extraction Project | ||
| Line 93: | Line 106: | ||
:* Extracting path of interest, and calculating FA along the path for group comparison. Presentation of previous results for 7 schizophrenics and 12 control subjects, can be found here: [[Media:NAMIC_AHM_Arcuate.ppt|Progress Report Presentation]]. | :* Extracting path of interest, and calculating FA along the path for group comparison. Presentation of previous results for 7 schizophrenics and 12 control subjects, can be found here: [[Media:NAMIC_AHM_Arcuate.ppt|Progress Report Presentation]]. | ||
:* Results presentation and paper submission. Abstract was submitted and accepted for presentation at World Biological Psychiatry Symposium in Venice, Italy. Paper is in preparation. | :* Results presentation and paper submission. Abstract was submitted and accepted for presentation at World Biological Psychiatry Symposium in Venice, Italy. Paper is in preparation. | ||
| + | |||
| + | | style="width:85%" | [[Image:STArcuate.jpg|thumb|right|200px|<font size=1>Figure 6: The arcuate fasciculus including seed, midpoint and target ROI's.</font>]] | ||
| + | |||
| + | |} | ||
| + | |||
| + | |||
| + | {|cellpadding="0" | ||
| + | |||
| + | | style="width:85%" | | ||
;* Semantic Network Connectivity Project | ;* Semantic Network Connectivity Project | ||
| Line 117: | Line 139: | ||
* Polina is the algorithm core contact | * Polina is the algorithm core contact | ||
* Brad is the engineering core contact | * Brad is the engineering core contact | ||
| + | |||
| + | | style="width:15%" | | ||
| + | |} | ||
===Schedule=== | ===Schedule=== | ||
Revision as of 19:38, 11 December 2008
Home < DBP2:Harvard:Brain Segmentation RoadmapBack to NA-MIC Collaborations, Harvard DBP 2
Stochastic Tractography for VCFS
Roadmap
The main goal of this project is to develop end-to-end application that would be used to characterize anatomical connectivity abnormalities in the brain of patients with velocardiofacial syndrome (VCFS), and to link this information with deficits in schizophrenia. This page describes the technology roadmap for stochastic tractography, using newly acquired 3T data, NAMIC tools and slicer 3.
Algorythm
|
|
- C-References
- Björnemo M, Brun A, Kikinis R, Westin CF. Regularized stochastic white matter tractography using diffusion tensor MRI. In Fifth International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI'02). Tokyo, Japan, 2002;435-442.
- Friman, O., Farneback, G., Westin CF. A Bayesian Approach for Stochastic White Matter Tractography. IEEE Transactions on Medical Imaging, Vol 25, No. 8, Aug. 2006
Module
Fill it in once its ready. Write exactly, step by step what is its functionality, adding snapshots:
- Loading DTI in Nrrd format
- eddy current distortion correction
- movement correction
- smoothing
- tensor estimation
- creating WM mask
- loading rois
- creating probability map
- visual inspection (looking at areas where mask does not exist, but FA exceeds 0.3)
- thresholding
- visualizing path, tracts
- getting numbers (thresholded FA, FA weighted by probability)
- Module documentation can be found here (replace with the new documentation, when its ready):
Work Accomplished
|
Work in Progress
|
We have started the project of investigating Arcuate Fasciculus using Stochastic Tractography (Figure 6). This structure is especially important in both VCFS and schizophrenia, as it connects language related areas (Brocka and Wernicke's), and is involved in language processing quite disturbed in schizophrenia patients. It also can not be reliably traced using deterministic tractography.
|
We use combination of fMRI and DTI data to define and characterize functional and anatomical connectivity within the semantic processing network in schizophrenia.
We have started collaboration with Department of Neurophysiology Max Planck Institute in Frankfurt (Anna Rotarska contact person). They have dataset containing DTI and resting state fMRI in schizophrenia, and want to use stochastic tractography to measure integrity of anatomical connections within the default network in schizophrenia. Their fMRI data has been co-registered with anatomical scans, and we are putting it into the DTI space. Once this is done, we will start creating tracts connecting fMRI ROIs. This data will be also used to test robustness of our module, since data was collected on a different scanner (3T Philips).
We are also working on a tractography comparison projectdataset, where we apply stochastic tractography to phantom, as well as test dataset. Staffing Plan
|
Schedule
- 10/2007 - Optimization of Stochastic Tractography algorythm for 1.5T data.
- 10/2007 - Algorythm testing on Santa Fe data set and diffusion phantom.
- 06/2008 - Optimization of Stochastic Tractography algorythm for 3T data.
- 11/2008 - Slicer 3 module prototype using python.
- 12/2008 - Slicer 3 module official release
- 12/2008 - Documentation and packaging for dissemination.
- 12/2008 - Arcuate Fasciculus results.
- 01/2009 - Arcuate Fasciuclus first draft of the paper.
- 05/2009 - Distortion correction and nonlinear registration added to the module
- 05/2009 - Symposium on tractography, including stochastic methods at World Biological Psychiatry Symposium in Venice, Italy.
- 05/2009 - Presentation of Arcuate Fasciculus findings at World Biological Psychiatry Symposium in Venice, Italy.
Team and Institute
- PI: Marek Kubicki (kubicki at bwh.harvard.edu)
- DBP2 Investigators: Sylvain Bouix, Yogesh Rathi, Julien de Siebenthal
- NA-MIC Engineering Contact: Brad Davis, Kitware
- NA-MIC Algorithms Contact: Polina Gollard, MIT
Publications
In print