Difference between revisions of "Engineering:GE"
| Line 63: | Line 63: | ||
Funded through the NCBC collaboration grant mechanism, this project brings together researchers from UNC and GE Research to develop deformable registration and MS lesion segmentation tools. [[NA-MIC_NCBC_Collaboration:Development_and_Dissemination_of_Robust_Brain_MRI_Measurement_Tools | More...]] | Funded through the NCBC collaboration grant mechanism, this project brings together researchers from UNC and GE Research to develop deformable registration and MS lesion segmentation tools. [[NA-MIC_NCBC_Collaboration:Development_and_Dissemination_of_Robust_Brain_MRI_Measurement_Tools | More...]] | ||
| + | |||
| + | |- | ||
| + | |||
| + | | align="center" | [[Image:SkullStrippingAndTissueClassification.png|220px]] | ||
| + | | | | ||
| + | |||
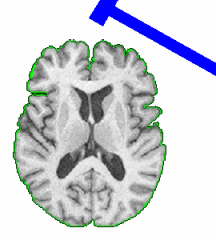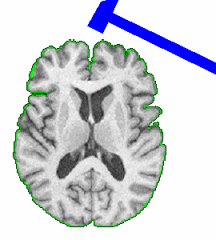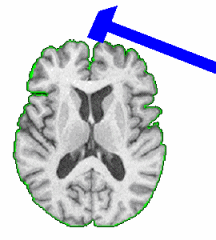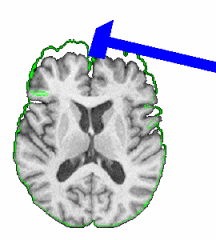
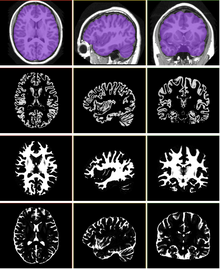
| + | == Skull Stripping and Tissue Classification == | ||
| + | |||
Revision as of 02:32, 15 May 2009
Home < Engineering:GEOverview (PI: Jim Miller)
GE is adding technology to ITK to support the needs of NA-MIC, developing a module plugin and scheduling architecture for Slicer3, developing a registration architecture, developing standard radiological review modes, and providing DICOM support for diffusion imaging.
GE Research Projects

|
Insight Toolkit (ITK)The Insight Segmentation and Registration Toolkit (ITK) is an open-source software toolkit for performing registration and segmentation. ITK is used as the principal library for image analysis and general image IO in the 3D Slicer. GE Research has extended ITK to support NA-MIC activities by providing probability distributions, tensor pixels, level tracing, and displacement transforms. |

|
Slicer3 Plugin ArchitectureThe 3D Slicer platform has been completely reworked through the joint efforts of the NA-MIC community and the BIRN, NAC, NCIGT, and other cooperative grants that leverage the common infrastructure. GE Research has created a simple plugin architecture that allows researchers to extend Slicer3's capabilities with plugins provided as executables, shared libraries, or python modules. See here for more information. We have recently extended the execution model to include nonlinear transformations (BSpline and Displacement Vectors) and have plans for CSV (Comma Separated Value) extensions soon. We have also revamped the Slicer3 build and installation configurations to allow plugins to be built outside of a Slicer build. See here and here for more information on building plugins outside of Slicer3. |
|
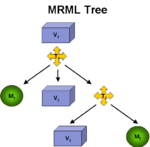
Registration ArchitectureA fundamental technology in Slicer is the scene graph or MRML scene used to describe the relationships between various sources of primary and derived data and annotation. Relating images from longitudinal or cross-sectional studies requires estimating the transformation between the image sets, a process referred to as registration. GE Research is providing an infrastructure for registration inside of Slicer3, leveraging existing registration algorithms from ITK, packaged as plugins that can be used inside or outside of Slicer3. The architecture includes IO mechanisms from transforms from files and plugins, transform representations in MRML, visualization through transforms, and managing the differences in coordinate frames between Slicer3 and ITK (see here and here). |
|
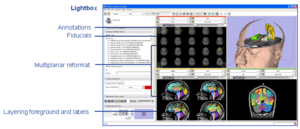
Radiological Review ModesClinical researchers expect image review workstations to provide a set of basic functionality. Handling DICOM as a source of image data is one example. Other examples include image multiplanar and oblique reformat, lightbox, comparisons, tracked cursors, ROI's, cine, and calipers for measurement. In conjunction with funding from NAC, GE Research is providing some of these base capabilities to Slicer3. More... |
|
DICOM Support for Diffusion ImagingDiffusion weighted MR images in DICOM format are not currently recognized by Slicer as DW-MR images due to many proprietary tags and data formats used by different vendors. We created a plugin module for converting DICOM series of diffusion weighted images into NRRD volumes where correct image information (diffusion weighting directions and b-values) are parsed from the DICOM header and stored in the appropriate fileds in NRRD header. The converter supports some of the common formats from GE, Siemens, and Philips. Once the list of supported formats is reletively complete, the DICOM support for diffusion weighted images will be moved to itkVTKArchetypeIO. |
|
Robust MRI Measurement ToolsFunded through the NCBC collaboration grant mechanism, this project brings together researchers from UNC and GE Research to develop deformable registration and MS lesion segmentation tools. More... |
|
Skull Stripping and Tissue Classification |