Difference between revisions of "Projects:RegistrationDocumentation:UseCaseInventoryNotes"
From NAMIC Wiki
m |
|||
| Line 5: | Line 5: | ||
<span style="color: rgb(50, 180, 0);">List of cases collected/inventoried thus far is here:</span> | <span style="color: rgb(50, 180, 0);">List of cases collected/inventoried thus far is here:</span> | ||
--> | --> | ||
| + | ==Categories & Formatting== | ||
| + | *Ideally we seek 2 sets for each use case to define a range of settings, i.e. one case that is average and works reasonably well, and one case that is more challenging. Both cases together then define a range. The two cases would be posted together, provided the difficult case can be solved with different settings and does not require a different approach. The latter case would then be better listed in a ''Troubleshooting'' category. | ||
| + | *The main categories will follow along the hierarchies outlined [[Projects:RegistrationDocumentation#Use_Case_Library|here]]. | ||
| + | *We seek to have all datasets in a single-volume file format, most likely '''.nii''' or '''.nrrd'''. This will greatly facilitate file management and case documentation and will also guarantee full anonymization. The NRRD format is defined [http://teem.sourceforge.net/nrrd/index.html here] and the NIFTI format is defined [http://nifti.nimh.nih.gov/nifti-1 here]. | ||
| + | *'''Anonymization''': Anonymized data is imperative. In a first pass posting single volume files only that do not contain subject-specific meta data is the safest. Relying on the provider/user to properly anonymize is likely insufficient. It's easy to overlook single DICOM fields. Defacing algorithms are avail. thru [[Mbirn:_Defacer_for_structural_MRI|MBIRN]] but will affect the result and require the mask to be avail. to the registration. | ||
| + | *Once formatting, anonymization and description are complete, listing here will be replaced with a download link. Temporarily listed are links to source databases, where avail. | ||
| + | *'''Speed''': include speed measurement with each case. Include note in tutorial that speed concerns are built into the default settings, i.e. if results do not look right on first run, there's many options to fine tune, hence the library concept | ||
| − | + | == Hierarchy / Organization == | |
[[Image:UseCaseHierarchySnapshot_108Levels.png|150px|right|108 Level Tree]] | [[Image:UseCaseHierarchySnapshot_108Levels.png|150px|right|108 Level Tree]] | ||
Revision as of 16:05, 10 November 2009
Home < Projects:RegistrationDocumentation:UseCaseInventoryNotesback to Use Case Library
back to Registration Main Page
Categories & Formatting
- Ideally we seek 2 sets for each use case to define a range of settings, i.e. one case that is average and works reasonably well, and one case that is more challenging. Both cases together then define a range. The two cases would be posted together, provided the difficult case can be solved with different settings and does not require a different approach. The latter case would then be better listed in a Troubleshooting category.
- The main categories will follow along the hierarchies outlined here.
- We seek to have all datasets in a single-volume file format, most likely .nii or .nrrd. This will greatly facilitate file management and case documentation and will also guarantee full anonymization. The NRRD format is defined here and the NIFTI format is defined here.
- Anonymization: Anonymized data is imperative. In a first pass posting single volume files only that do not contain subject-specific meta data is the safest. Relying on the provider/user to properly anonymize is likely insufficient. It's easy to overlook single DICOM fields. Defacing algorithms are avail. thru MBIRN but will affect the result and require the mask to be avail. to the registration.
- Once formatting, anonymization and description are complete, listing here will be replaced with a download link. Temporarily listed are links to source databases, where avail.
- Speed: include speed measurement with each case. Include note in tutorial that speed concerns are built into the default settings, i.e. if results do not look right on first run, there's many options to fine tune, hence the library concept
Hierarchy / Organization
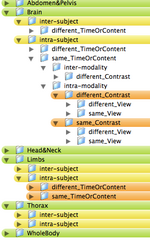
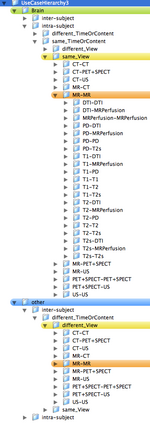
- The registration case library will contain the most common scenarios encountered for Slicer Registration. Each case will contain a dataset, a parameter set, a guided tutorial with example result. The hierarchy will be represented graphically as a tree and as a list with links to the abovementioned data. The library will be as exhaustive as possible, containin brain and non-brain, different modalities (MRI, CT, PET/SPECT).
- considered is also a list of troubleshooting cases, i.e. a list of the most common sources of registration failure, again complete with dataset, tutorial and remedy.
- the organization of a use case hierarchy can be accomplished in several ways. The main categories are listed below. Not all combinations exist or are equally common occurrences. For example, a different-timepoint/contrast distinction is not necessary for inter-subject registration.
- ORGAN: Brain, Head&Neck, Thorax, Abdomen&Pelvis, Limbs, WholeBody
- SUBJECT: intra-subject, inter-subject
- MODALITY: intra-modality, inter-modality
- CONTRAST: same_Contrast, different_Contrast
- MR, CT, PET+SPECT, US
- T1, T2, PD, T2s, DTI, MRPerfusion
- CONTENT: same_TimeOrContent, different_TimeOrContent
- VIEW: same_View, different_View
- Depending on grouping this leads to trees with 80-1080 entries. Two example hierarchies (directory trees) are here: 108 Level Tree , 360 Level Tree
- the VIEW distinction arises from commonly occurring problems of co-registering sets acquired with different slice orientation. The basics of dealing with axis orientation is already integrated into the algorithm, but further details remain: basically different_view implies a possibly high voxel size ratio (i.e. different voxel sizes if one image acquired axial the other coronal). Actions/Parameter settings are with multi-resolution settings, blurring and special checking of the axis orientation (vox2ras) meta-data. If, for example,voxel size ratio is close to [1 1 1] we need not do anything different other than resample based on vox2ras.
- No separate distinction for Atlas Registration, i.e. the case where the volume of interest is not the one used for calculating the transform.
- User-populated content The exploding # of combinations makes a successful top-down approach difficult. More effective to organize bottom-up, i.e. have user invited content, which we organize into a growing tree. We can make a call to the user community for submitting datasets and registration problems. This call should be in the "Register Images" Documentation as well as the "Help&Acknowledgement" tab of the "Register Images" module. e.g. Post a link on the documentation pages and module info tabs: Having trouble with registration? Try our service. We will consider all cases, provided that you have already tried what we do offer on the library and that did not work. If it didn't then that speaks for having your case added.