Difference between revisions of "Projects:RegistrationLibrary:RegLib C10"
From NAMIC Wiki
| Line 12: | Line 12: | ||
[[Image:Button_red_fixed.jpg|20px|lleft]] this indicates the reference image that is fixed and does not move. All other images are aligned into this space and resolution<br> | [[Image:Button_red_fixed.jpg|20px|lleft]] this indicates the reference image that is fixed and does not move. All other images are aligned into this space and resolution<br> | ||
[[Image:Button_green_moving.jpg|20px|lleft]] this indicates the moving image that determines the registration transform. <br> | [[Image:Button_green_moving.jpg|20px|lleft]] this indicates the moving image that determines the registration transform. <br> | ||
| − | + | </small></small> | |
|- | |- | ||
|[[Image:Button_red_fixed.jpg|40px|lleft]] Target Brain | |[[Image:Button_red_fixed.jpg|40px|lleft]] Target Brain | ||
| | | | ||
| − | |[[Image:Button_green_moving.jpg|40px|lleft]] | + | |[[Image:Button_green_moving.jpg|40px|lleft]] Probabilistic Atlas (T1w) |
|- | |- | ||
| − | |0. | + | |0.9375 x 0.9375 x 1.5 mm axial <br> 256 x 256 x 124<br>T1w, PA |
| | | | ||
| − | | | + | |0.9375 x 0.9375 x 1.5 mm axial <br> 256 x 256 x 124<br>T1w, PA |
|} | |} | ||
| Line 31: | Line 31: | ||
*[[Image:Button_red_fixed_white.jpg|20px]]reference/fixed : T1w axial, 1mm resolution in plane, 3mm slices | *[[Image:Button_red_fixed_white.jpg|20px]]reference/fixed : T1w axial, 1mm resolution in plane, 3mm slices | ||
*[[Image:Button_green_moving_white.jpg|20px]] moving: Probabilistic Tissue atlas, | *[[Image:Button_green_moving_white.jpg|20px]] moving: Probabilistic Tissue atlas, | ||
| − | |||
=== Methods === | === Methods === | ||
#build brain mask for fixed image using '''Skull Stripping''' module. Settings: 100 iterations, 20 subdivisions. New Volume: ''RegLib_C10_MRI_AtlasSegmentation_fixed_mask'' | #build brain mask for fixed image using '''Skull Stripping''' module. Settings: 100 iterations, 20 subdivisions. New Volume: ''RegLib_C10_MRI_AtlasSegmentation_fixed_mask'' | ||
| Line 50: | Line 49: | ||
::*Affine Max Iteration: 80 | ::*Affine Max Iteration: 80 | ||
::*Affine Sampling Ratio: 0.05 | ::*Affine Sampling Ratio: 0.05 | ||
| − | + | #(alternatively automated Affine Registration: '''Register Images Multires''' (Slicer 3.5) also produces good results | |
# run '"Deformable B-spline Registration'" module. Settings: | # run '"Deformable B-spline Registration'" module. Settings: | ||
::*Grid Size: 5 | ::*Grid Size: 5 | ||
| Line 56: | Line 55: | ||
::*Spatial Samples: 50000, | ::*Spatial Samples: 50000, | ||
::*initial transform: ''RegLib_C10_MRI_AtlasSegmentation_Xform_Affine_wmsk'' | ::*initial transform: ''RegLib_C10_MRI_AtlasSegmentation_Xform_Affine_wmsk'' | ||
| − | |||
| − | |||
| − | |||
| − | |||
=== Registration Results=== | === Registration Results=== | ||
| Line 88: | Line 83: | ||
=== Discussion: Registration Challenges === | === Discussion: Registration Challenges === | ||
| − | *Because the atlas represents a | + | *Because the atlas represents a probabilistic image (i.e. contains blurring from combining multiple subjects), its contrast differs significantly from the target image. |
| − | *The | + | *The atlas has strong rotational misalignment that can cause difficulty for automated affine registration. |
*The two images represent different anatomies, a non-rigid registration is required | *The two images represent different anatomies, a non-rigid registration is required | ||
| Line 95: | Line 90: | ||
*Because of the strong differences in image contrast, Mutual Information is recommended as the most robust metric. | *Because of the strong differences in image contrast, Mutual Information is recommended as the most robust metric. | ||
*masking (skull stripping) is highly recommended to obtain good results. | *masking (skull stripping) is highly recommended to obtain good results. | ||
| − | *because speed is not that critical, we increase the sampling rate | + | *because speed is not that critical, we increase the sampling rate for both affine and BSpline registration |
*we also expect larger differences in scale & distortion than with regular structural scans: so we significantly (2x-3x) increase the expected values for scale and skew from the defaults. | *we also expect larger differences in scale & distortion than with regular structural scans: so we significantly (2x-3x) increase the expected values for scale and skew from the defaults. | ||
*a good affine alignment is important before proceeding to non-rigid alignment to further correct for distortions. | *a good affine alignment is important before proceeding to non-rigid alignment to further correct for distortions. | ||
=== Acknowledgments === | === Acknowledgments === | ||
| + | *dataset provided by Killian Pohl, Ph.D. | ||
Revision as of 15:48, 17 February 2010
Home < Projects:RegistrationLibrary:RegLib C10Back to ARRA main page
Back to Registration main page
Back to Registration Use-case Inventory
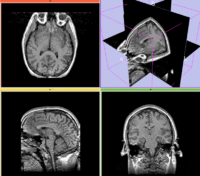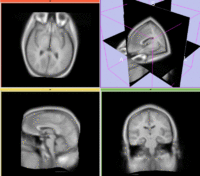
Slicer Registration Library Exampe #10: Co-registration of probabilistic tissue atlas for subsequent EM segmentation
Objective / Background
This is an example of sparse atlas co-registration. Not all atlases have an associated reference image that can be used for registration. Because the atlas represents a map of a particular tissue class probability, its contrast differs significantly from the target image.
Keywords
MRI, brain, head, inter-subject, probabilistic atlas, atlas-based segmentation
Input Data
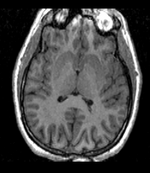
 reference/fixed : T1w axial, 1mm resolution in plane, 3mm slices
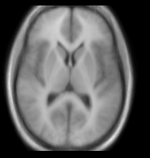
reference/fixed : T1w axial, 1mm resolution in plane, 3mm slices moving: Probabilistic Tissue atlas,
moving: Probabilistic Tissue atlas,
Methods
- build brain mask for fixed image using Skull Stripping module. Settings: 100 iterations, 20 subdivisions. New Volume: RegLib_C10_MRI_AtlasSegmentation_fixed_mask
- manually edit brain mask with Editor. required manual fix at frontal and occipital lobe
- run Register Images , Settings:
- Fixed Image:
- Moving Image:
- Resample Image:
- Load Transform:
- Save Transform: RegLib_C10_MRI_AtlasSegmentation_Xform_Affine_wmsk
- Initialization: Centers of Mass,
- Registration: PipelineAffine
- Expected offset: 10
- Expected Rotation: 0.2
- Expected Scale: 0.1
- Expected Skew: 0.05
- Fixed Image Mask: RegLib_C10_MRI_AtlasSegmentation_fixed_mask
- Affine Max Iteration: 80
- Affine Sampling Ratio: 0.05
- (alternatively automated Affine Registration: Register Images Multires (Slicer 3.5) also produces good results
- run '"Deformable B-spline Registration'" module. Settings:
- Grid Size: 5
- Histogram Bins: 50,
- Spatial Samples: 50000,
- initial transform: RegLib_C10_MRI_AtlasSegmentation_Xform_Affine_wmsk
Registration Results
Download
- download entire package (Data,Tutorial, Solution, zip file 14 MB)
- Presets
- Tutorial only
- Image Data only
Discussion: Registration Challenges
- Because the atlas represents a probabilistic image (i.e. contains blurring from combining multiple subjects), its contrast differs significantly from the target image.
- The atlas has strong rotational misalignment that can cause difficulty for automated affine registration.
- The two images represent different anatomies, a non-rigid registration is required
Discussion: Key Strategies
- Because of the strong differences in image contrast, Mutual Information is recommended as the most robust metric.
- masking (skull stripping) is highly recommended to obtain good results.
- because speed is not that critical, we increase the sampling rate for both affine and BSpline registration
- we also expect larger differences in scale & distortion than with regular structural scans: so we significantly (2x-3x) increase the expected values for scale and skew from the defaults.
- a good affine alignment is important before proceeding to non-rigid alignment to further correct for distortions.
Acknowledgments
- dataset provided by Killian Pohl, Ph.D.