Difference between revisions of "Projects:RegistrationLibrary:RegLib C09"
From NAMIC Wiki
| Line 3: | Line 3: | ||
[[Projects:RegistrationDocumentation:UseCaseInventory|Back to Registration Use-case Inventory]] <br> | [[Projects:RegistrationDocumentation:UseCaseInventory|Back to Registration Use-case Inventory]] <br> | ||
| − | ==Slicer Registration Library | + | == <small>v3.6.3</small> [[Image:Slicer3-6Announcement-v1.png|150px]] Slicer Registration Library Case 09: Functional MRI aligned with structural reference MRI == |
| − | + | === Input === | |
| − | + | {| style="color:#bbbbbb; " cellpadding="10" cellspacing="0" border="0" | |
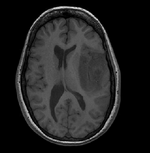
| − | {| style="color:#bbbbbb | + | |[[Image:RegLib_C09_Thumb1.png|150px|lleft|this is the fixed T1 reference image.]] |
| − | |[[Image: | + | |[[Image:RegArrow_NonRigid.png|100px|lleft]] |
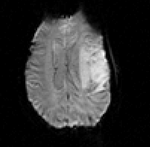
| − | |[[Image: | + | |[[Image:RegLib_C09_Thumb2.png|150px|left|this is the fMRI baseline image, to be registered to the T1]] |
| − | |[[Image: | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | |Target Anatomical Ref. |
| | | | ||
| − | | | + | |fMRI |
|} | |} | ||
| + | === Modules === | ||
| + | *'''Slicer 3.6.1 recommended modules: [http://www.slicer.org/slicerWiki/index.php/Modules:BRAINSFit BrainsFit]''' | ||
===Objective / Background === | ===Objective / Background === | ||
This is a typical example of fMRI pre-processing. Goal is to align the fMRI image with a structural scan that provides accuracte anatomical reference. The fMRI contains acquisition-related distortion and low contrast to discern much anatomical detail. We also have pathology (stroke) with variable contrast across different MRI protocols. | This is a typical example of fMRI pre-processing. Goal is to align the fMRI image with a structural scan that provides accuracte anatomical reference. The fMRI contains acquisition-related distortion and low contrast to discern much anatomical detail. We also have pathology (stroke) with variable contrast across different MRI protocols. | ||
| + | |||
=== Keywords === | === Keywords === | ||
MRI, brain, head, intra-subject, fMRI | MRI, brain, head, intra-subject, fMRI | ||
===Input Data=== | ===Input Data=== | ||
| − | * | + | *reference/fixed : T1 0.5 x 0.5 x 1 mm , 512 x 512 x 176 |
| − | * | + | *moving: fMRI sequence of motor task (right hand clench) 2 x 2 x 4 mm, 128 x 128 x 19 |
| − | |||
| − | |||
| − | |||
===Download === | ===Download === | ||
| − | *'''[[Media: | + | *'''[[Media:RegLib_C09_Data.zip|download example data <small> (Data,Presets, Solution, zip file 60 MB) </small>]]''' |
| − | * | + | *'''[[Media:RegLib_C09_Presets.mrml|download parameter preset file <small> ( MRML file 12 kB) </small>]]''' |
| − | + | :[[Projects:RegistrationDocumentation:ParameterPresetsTutorial|Link to User Guide: How to Load/Save Registration Parameter Presets]] | |
| − | |||
| − | |||
| − | |||
| − | [[Projects:RegistrationDocumentation:ParameterPresetsTutorial|Link to User Guide: How to Load/Save Registration Parameter Presets]] | ||
=== Discussion: Registration Challenges === | === Discussion: Registration Challenges === | ||
| Line 54: | Line 40: | ||
=== Discussion: Key Strategies === | === Discussion: Key Strategies === | ||
| − | * | + | *this example manages to lock onto the target w/o masking, despite the FOV clipping. For similar cases it is likely that masking of the brain in both images may be necessary to obtain a good match. |
| − | + | *a good rigid alignment is important before proceeding to non-rigid alignment to further correct for distortions. Affine is not recommended w/o masking, because the FOV differences would incur large scaling distortions. | |
| − | + | *The nonrigid portion should be constrained to a max. deformation, e.g. 3mm | |
| − | |||
| − | *a good | ||
| − | === | + | === Methods === |
| + | *place SPGR in background and fMRI in foreground. Review initial misalignment. | ||
| + | *if there is little or no initial overlap, go to the [http://www.slicer.org/slicerWiki/index.php/Modules:Volumes-Documentation-3.6 Volumes module], select the ''Info'' tab and click the ''Volume'' button. Repeat for both images. This should roughly center the volumes in the same physical space. Alternatively, when loading the data, consider checking the box for "Centered'' in the "Open File" dialog | ||
| + | *'''Phase 1: BRAINSfit rigid w/o masking''' | ||
| + | #open the Registration / [http://www.slicer.org/slicerWiki/index.php/Modules:BRAINSFit BRAINSfit module] | ||
| + | ##Select Preset "Xf0_Rigid" or set the parameters as shown below: | ||
| + | ##fixed image: SPGR; moving image: fMRI | ||
| + | ##Registration phases: | ||
| + | ###Initialize check: useGeometryAlign; check: Rigid | ||
| + | ##Output: under ''Slicer BSpline Transform'', select "create new" and rename to "Xf3_BFit_unmasked" | ||
| + | ##Output: under ''Output Image Volume'', select "create new" and rename to "moving_Xf3"ed'' checkbox | ||
| + | ##Output Image Pixel Type: check box for "short" | ||
| + | ##Registration Parameters: increase ''Number of Samples'' to 200,000 | ||
| + | ##Number of Grid Subdivisions: 5,5,5 | ||
| + | ##leave rest at defaults | ||
| + | ##click: ''Apply'' | ||
| + | *'''Version 2: Expert Automated + Fast BSpline Modules incl. masking''' | ||
| + | #build brain mask for the fixed image only: | ||
| + | ##open '''Skull Stripping''' module. | ||
| + | ##Settings: 100 iterations, 20 subdivisions. | ||
| + | ##Ouput: create new volum, rename to "fixed_mask" | ||
| + | ##Click: ''Apply'' | ||
| + | ##manually edit brain mask with '''Editor''' module. required manual fix at frontal and occipital lobe | ||
| + | #open ''Expert Automated Registration'' module | ||
| + | ##Settings: Fixed Image: fixed, moving image: moving | ||
| + | ##Save Transform: ''Xf1_Affine_wmsk'' | ||
| + | ##Initialization: Centers of Mass, Registration: PipelineAffine | ||
| + | ##Expected offset: 10 , Expected Rotation: 0.2 | ||
| + | ##Expected Scale: 0.1 , Expected Skew: 0.05 | ||
| + | ##Fixed Image Mask: "fixed_mask" | ||
| + | ##Affine Max Iteration: 80 , Affine Sampling Ratio: 0.05 | ||
| + | #(alternatively automated Affine Registration: '''Register Images Multires''' also produces good results | ||
| + | # run '''Fast Deformable B-spline Registration''' module. Settings: | ||
| + | ##Iterations: 20 , Grid Size: 9 | ||
| + | ##fixed image: fixed; moving image: moving | ||
| + | ##Histogram Bins: 50, Spatial Samples: 50000 | ||
| + | ## initial transform: "Xf1_Affine_wmsk" | ||
| + | ##Output Transform: Xf2_BSpline1 | ||
| + | ##Output Volume: create new, rename to "moving_Xf2" | ||
| + | |||
| + | === Registration Results=== | ||
| + | [[Image:RegLib_C10_unreg_AnimGif.gif|300px|unregistered]] unregistered | ||
| + | [[Image:RegLib_C10_BSpline_AnimGif.gif|300px|after BSpline non-rigid registration]] afterAffine+ fast BSpline non-rigid registration<br><br> | ||
| + | [[Image:RegLib_C10_BFit_AGif.gif|400px|after BRAINSfit affine+ non-rigid registration]] after BRAINSfit affine+ non-rigid registration<br> | ||
Revision as of 20:00, 26 August 2011
Home < Projects:RegistrationLibrary:RegLib C09Back to ARRA main page
Back to Registration main page
Back to Registration Use-case Inventory
Contents
v3.6.3  Slicer Registration Library Case 09: Functional MRI aligned with structural reference MRI
Slicer Registration Library Case 09: Functional MRI aligned with structural reference MRI
Input
|
|
|
| Target Anatomical Ref. | fMRI |
Modules
- Slicer 3.6.1 recommended modules: BrainsFit
Objective / Background
This is a typical example of fMRI pre-processing. Goal is to align the fMRI image with a structural scan that provides accuracte anatomical reference. The fMRI contains acquisition-related distortion and low contrast to discern much anatomical detail. We also have pathology (stroke) with variable contrast across different MRI protocols.
Keywords
MRI, brain, head, intra-subject, fMRI
Input Data
- reference/fixed : T1 0.5 x 0.5 x 1 mm , 512 x 512 x 176
- moving: fMRI sequence of motor task (right hand clench) 2 x 2 x 4 mm, 128 x 128 x 19
Download
- download example data (Data,Presets, Solution, zip file 60 MB)
- download parameter preset file ( MRML file 12 kB)
Discussion: Registration Challenges
- the fMRI contains acquisition-related distortions that can make automated registration difficult.
- the fMRI contains low tissue contrast, making automated intensity-based registration difficult.
- the two images often have strong differences in voxel sizes and voxel anisotropy. If the orientation of the highest resolution is not the same in both images, finding a good match can be difficult.
- there may be widespread and extensive pathology (e.g stroke, tumor) that might affect the registration if its contrast is different in the baseline and structural reference scan
Discussion: Key Strategies
- this example manages to lock onto the target w/o masking, despite the FOV clipping. For similar cases it is likely that masking of the brain in both images may be necessary to obtain a good match.
- a good rigid alignment is important before proceeding to non-rigid alignment to further correct for distortions. Affine is not recommended w/o masking, because the FOV differences would incur large scaling distortions.
- The nonrigid portion should be constrained to a max. deformation, e.g. 3mm
Methods
- place SPGR in background and fMRI in foreground. Review initial misalignment.
- if there is little or no initial overlap, go to the Volumes module, select the Info tab and click the Volume button. Repeat for both images. This should roughly center the volumes in the same physical space. Alternatively, when loading the data, consider checking the box for "Centered in the "Open File" dialog
- Phase 1: BRAINSfit rigid w/o masking
- open the Registration / BRAINSfit module
- Select Preset "Xf0_Rigid" or set the parameters as shown below:
- fixed image: SPGR; moving image: fMRI
- Registration phases:
- Initialize check: useGeometryAlign; check: Rigid
- Output: under Slicer BSpline Transform, select "create new" and rename to "Xf3_BFit_unmasked"
- Output: under Output Image Volume, select "create new" and rename to "moving_Xf3"ed checkbox
- Output Image Pixel Type: check box for "short"
- Registration Parameters: increase Number of Samples to 200,000
- Number of Grid Subdivisions: 5,5,5
- leave rest at defaults
- click: Apply
- Version 2: Expert Automated + Fast BSpline Modules incl. masking
- build brain mask for the fixed image only:
- open Skull Stripping module.
- Settings: 100 iterations, 20 subdivisions.
- Ouput: create new volum, rename to "fixed_mask"
- Click: Apply
- manually edit brain mask with Editor module. required manual fix at frontal and occipital lobe
- open Expert Automated Registration module
- Settings: Fixed Image: fixed, moving image: moving
- Save Transform: Xf1_Affine_wmsk
- Initialization: Centers of Mass, Registration: PipelineAffine
- Expected offset: 10 , Expected Rotation: 0.2
- Expected Scale: 0.1 , Expected Skew: 0.05
- Fixed Image Mask: "fixed_mask"
- Affine Max Iteration: 80 , Affine Sampling Ratio: 0.05
- (alternatively automated Affine Registration: Register Images Multires also produces good results
- run Fast Deformable B-spline Registration module. Settings:
- Iterations: 20 , Grid Size: 9
- fixed image: fixed; moving image: moving
- Histogram Bins: 50, Spatial Samples: 50000
- initial transform: "Xf1_Affine_wmsk"
- Output Transform: Xf2_BSpline1
- Output Volume: create new, rename to "moving_Xf2"
Registration Results
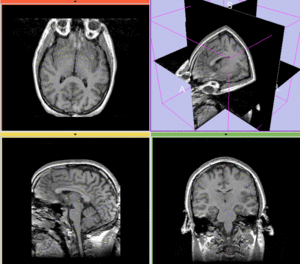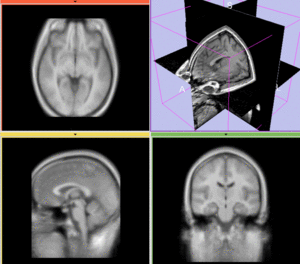 unregistered
unregistered
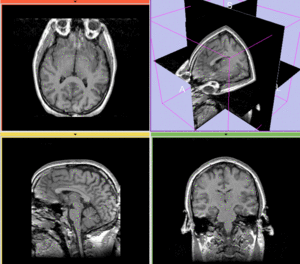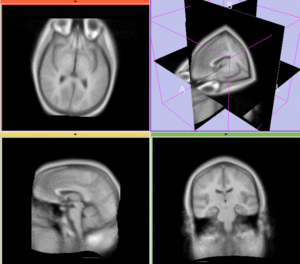 afterAffine+ fast BSpline non-rigid registration
afterAffine+ fast BSpline non-rigid registration
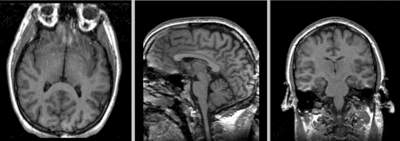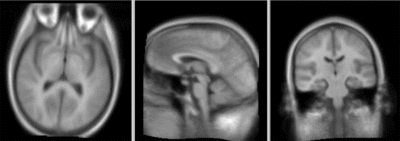 after BRAINSfit affine+ non-rigid registration
after BRAINSfit affine+ non-rigid registration