Difference between revisions of "Projects:RegistrationLibrary:RegLib C33"
From NAMIC Wiki
| Line 169: | Line 169: | ||
=== Acknowledgments === | === Acknowledgments === | ||
| + | This case is the same data as [http://central.xnat.org/app/action/DisplayItemAction/search_element/xnat%3AmrSessionData/search_field/xnat%3AmrSessionData.ID/search_value/CENTRAL_E00893/popup/false/project/IGT_FMRI_NEURO '''case 40''' ] from the fMRI neurosurgery data set on central.xnat.org | ||
Revision as of 15:36, 29 November 2010
Home < Projects:RegistrationLibrary:RegLib C33Back to ARRA main page
Back to Registration main page
Back to Registration Use-case Inventory
v3.6.1  Slicer Registration Library Exampe #33: Diffusion Weighted Image Volume: align with structural reference MRI
Slicer Registration Library Exampe #33: Diffusion Weighted Image Volume: align with structural reference MRI
Input
|
|
|
|
|
|
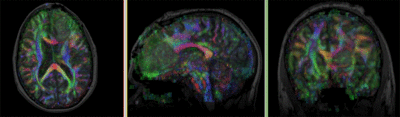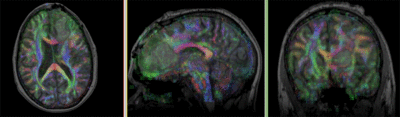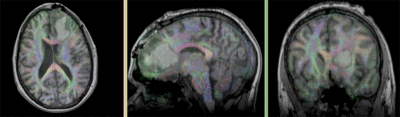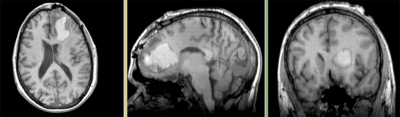
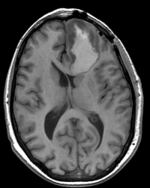
| fixed image/target T1 |
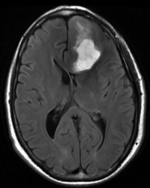
moving image FLAIR |
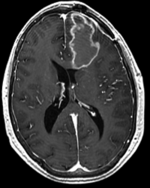
moving image 2a T1Gd |
moving image 2b DTI tensor |
Modules
- Slicer 3.6.1 recommended modules: BrainsFit
Objective / Background
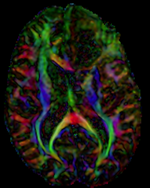
This is a typical example of DTI processing. Goal is to align the DTI image with a structural scan that provides accuracte anatomical reference. The DTI contains acquisition-related distortion and insufficient contrast to discern anatomical detail. For treatment planning and evaluation, location of functionally critical fiber tracts relative to the pathology is sought.
Keywords
MRI, brain, head, intra-subject, DTI, DWI
Input Data
- reference/fixed : T1 axial, 0.5 x 0.5 x 1 , 512 x 512 x 176
- moving : FLAIR axial
- moving : T1Gd axial
- moving: Tensor data of DWI volume, 32 directions, 2.6 mm slice thickness
Overall Strategy
- DWI -> DTI, obtain mask +baseline
- Subsample T1 to isotropic 1x1x1 mm -> T1_sub
- Register DTI_baseline to T1_sub (affine+nonrigid) w/o masking
- Resample DTI_mask with above transform to obtain mask for T1_sub
- Re-register DTI_baseline to T1_sub (affine+nonrigid) with masking
- Resample DTI with result nonrigid transform
- Resample DTI_mask with above nonrigid transform and FLAIR as reference -> yields mask for FLAIR
- Resample DTI_mask with above nonrigid transform and T1Gd as reference -> yields mask for T1Gd
- Register FLAIR and T1Gd to T1_sub (affine only) with masking
Procedures
- Phase I: Load & Convert DICOM
- run Dicom2NRRD converter for the DWI Dicom set. Output file: DWI.nrrd
- load DWI.nrrd
- load T1, FLAIR and T1Gd images via AddVolume...
- Subsample T1: The T1 is 512 x 512 x 176 (0.5 x 0.5 x 1) resolution, which is too large as a reference to sample the DTI into. So we generate a smaller, isotropic 1x1x1 version of the T1 that can serve as reference:
- Open Filtering/Resample Scalar Volume module
- Input: T1, Output: new volume, rename to T1_sub
- interpolation: Hamming
- Spacing: 1,1,1
- Phase II: Check initial Alignment and anisotropy
- large initial misalignment and high voxel anisotropy can cause problems when registering and resampling the DTI. In such cases it is advisable to resample the DWI first into a more isotropic space and, if necessary, initially realign to a similar orientation as the T1 reference, 'before converting to DTI. In this example DWI is sufficiently aligned and has voxel dimensions of 1 x 1 x 2.6, which is borderline. Anisotropy ratios greater than 1:3 tend to benefit from initial resampling. In this case we proceed without.
- 'Phase III: convert DWI -> DTI
- go to module: Diffusion / Utilities / Diffusion Tensor Estimation
- input: DWI, output : DTI, DTI_base and DTI_mask
- Input DWI Volume: DWI, Output DTI Volume: create new, rename to "DTI"
- Output Baseline: create new, rename to "DTI_base"
- Otsu Threshold Mask: create new, rename to "DTI_iso_mask"
- check boxes for Remove Islands and Apply Mask
- leave defaults
- Phase IVa: Register DTI-T1 (unmasked)
- open Registration : BrainsFit module (presets: Xf1_DTI-T1_unmasked
- Registration Phases:
- set T1_sub as fixed and DTI_base as moving image
- select/check Include Affine registration phase
- select/check Include BSpline registration phase
- Output Settings:
- select a new transform "Slicer BSpline Transform", rename to "Xf1_DTI-T1_unmasked"
- select a new volume "Output Image Volume, rename to "DTI_base_Xf1"
- Registration Parameters: increase Number Of Samples to 200,000
- Registration Parameters: set Number Of Grid Subdivisions to 5,5,3
- Leave all other settings at default
- click: Apply; (runtime < 1 min 10 sec. on MacPro Quad Core 2.4Ghz)
- result: the absence of the skull in the DTI causes significant and incorrect stretch of image in the I-S direction.
- we repeat with mask, using the above estimate to produce a mask for the T1
- Phase IVb: Build mask for T1: resample DTI_mask
- go to Filtering/Resample Scalar Vector DWI Volume
- Use provided presets or set the following:
- input: DTI_mask
- reference: T1_sub
- output: create new, rename to T1_sub_mask
- Transform node: Xf1_DTI-T1_unmasked
- Transforms order: check output-to-input
- interpolation type: check "nn"
- click: Apply
- Go to Volumes module
- select new "T1_sub_mask: volume, go to Info tab
- check the labelmap checkbox
- Phase IVc: Register DTI-T1 (masked)
- open Registration : BrainsFit module (presets: Xf1_DTI-T1_masked)
- settings as in Phase IVa above, except:
- select a new transform "Slicer BSpline Transform", rename to "Xf2_DTI-T1_masked"
- select a new volume "Output Image Volume, rename to "DTI_base_Xf2"
- Mask Processing Mode: check "ROI"
- Input Fixed Mask: T1_mask
- Input Moving Mask: DTI_mask
- Apply. Runtime 45 sec.
- go to Volumes module to adjust window&level for new result "DTI_base_Xf2"
- alignment is now much improved
- settings as in Phase IVa above, except:
- Phase V: Resample DTI
- Open the Resample DTI Volume module (found under: All Modules)
- Input Volume: select DTI
- Output Volume: select New DTI Volume, rename to DTI_Xf2
- Reference Volume: select T1_sub
- Transform Parameters: select transform "Xf2_DTI-T1_masked
- check box: output-to-input
- Leave all other settings at defaults
- Click Apply; runtime ~ 3 min.
- Phase VIa: Register FLAIR & T1 Gd: obtain masks
- we seek affine alignment only for FLAIR and T1Gd. As above, better results are achieved when masking non-brain parenchyma. To obtain masks for the FLAIR and T1Gd we proceed as above by resampling the DT_mask with the Xf2_DTI-T1 transform and the respective image as reference.
- go to Filtering/Resample Scalar Vector DWI Volume
- input: DTI_mask
- reference: FLAIR
- output: create new, rename to FLAIR_mask
- Transform node: Xf2_DTI-T1_masked
- Transforms order: check output-to-input
- interpolation type: check "nn"
- click: Apply
- Go to Volumes module
- select new "FLAIR_mask: volume, go to Info tab
- check the labelmap checkbox
- Phase VIb: Mask for T1Gd
- ditto: we seek affine alignment only for T1Gd. Identical procedure as for FLAIR above:
- go to Filtering/Resample Scalar Vector DWI Volume
- input: DTI_mask
- reference: T1Gd
- output: create new, rename to T1Gd_mask
- Transform node: Xf2_DTI-T1_masked
- Transforms order: check output-to-input
- interpolation type: check "nn"
- click: Apply
- Go to Volumes module
- select new "T1Gd_mask: volume, go to Info tab
- check the labelmap checkbox
- Phase VIc: Register FLAIR & T1Gd
- open Registration : BrainsFit module (presets: Xf4_... and Xf5_...)
- Registration Phase: check box for "Affine" only
- select a new linear transform "Slicer Linear Transform", rename to "Xf4_FLAIR-T1_masked"
- Mask Processing Mode: check "ROI"
- Input Fixed Mask: T1_mask
- Input Moving Mask: FLAIR_mask
- Apply. Runtime ~45 sec.
- Registration Phase: check box for "Affine" only
- ditto for T1Gd
Registration Results
Download
- Data
- RegLib_C33_Data DWI, DTI, T1, FLAIR, Presets & solution transforms (NRRD files, zip file 174 MB)
- RegLib_C33_Data_noDWI subset of the above, contains DTI but w/o DWI and resampled DTI (NRRD files, zip file 86 MB)
- RegLib_C33_Data Transforms & Presets only contains only result transforms and parameter presets (NRRD files, zip file 30 kB)
- Presets
Discussion: Registration Challenges
- The DTI contains acquisition-related distortions (commonly EPI acquisitions) that can make automated registration difficult.
Discussion: Key Strategies
Acknowledgments
This case is the same data as case 40 from the fMRI neurosurgery data set on central.xnat.org