Difference between revisions of "Projects:RegistrationLibrary:RegLib C12"
From NAMIC Wiki
| Line 3: | Line 3: | ||
[[Projects:RegistrationDocumentation:UseCaseInventory|Back to Registration Use-case Inventory]] <br> | [[Projects:RegistrationDocumentation:UseCaseInventory|Back to Registration Use-case Inventory]] <br> | ||
| − | ==Slicer Registration | + | ==<small>v3.6.3</small> [[Image:Slicer3-6Announcement-v1.png|150px]] Slicer Registration Library Case #12: Liver Tumor Cryoablation == |
| − | {| style="color:#bbbbbb | + | === Input === |
| − | |[[Image: | + | {| style="color:#bbbbbb; " cellpadding="10" cellspacing="0" border="0" |
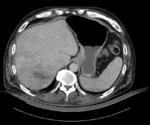
| − | |[[Image: | + | |[[Image:RegLib_C12_LiverTumor_CTpre.png|150px|lleft|this is the intra-op CT reference image. All images are aligned into this space]] |
| − | |[[Image: | + | |[[Image:RegArrow_NonRigid.png|100px|lleft]] |
| − | |[[Image: | + | |[[Image:RegLib_C12_LiverTumor_CTcontrast-pre.png|150px|lleft|this is an intermediate pre-op CT, used as reference to match the MRI]] |
| − | | | + | |[[Image:RegArrow_NonRigid.png|100px|lleft]] |
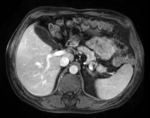
| − | [[Image: | + | |[[Image:RegLib_C12_LiverTumor_MRI.png|150px|lleft|this is the pre-op MRI we seek to align with the intra-op CT]] |
| − | |||
|- | |- | ||
| − | | | + | |fixed image/target |
| | | | ||
| − | | | + | |intermediate ref. image |
| − | |||
| − | |||
| − | |||
| | | | ||
| − | | | + | |moving image |
| − | |||
|} | |} | ||
| + | |||
| + | === Modules === | ||
| + | *'''Slicer 3.6.3 recommended modules: [http://www.slicer.org/slicerWiki/index.php/Modules:BRAINSFit BrainsFit]''' | ||
| + | |||
===Objective / Background === | ===Objective / Background === | ||
| − | We seek to align | + | We seek to align a pre-operative MRI with the intra-operative CT to enhance tumor visibility for surgical guidance. |
=== Keywords === | === Keywords === | ||
| Line 31: | Line 30: | ||
===Input Data=== | ===Input Data=== | ||
| − | * | + | *reference/fixed : pr-op CT, 0.95 x 0.95 x 5 mm voxel size |
| − | * | + | *moving: intra-op MRI, 0.78 x 0.78 x 2.5 mm axial, |
| + | |||
| + | === Notes / Overall Strategy === | ||
| + | *because the torso is already well registered and does not move across the sequence, a global affine/rigid registration is of no use. Instead we seek to assess the internal displacements caused across the breathing cycle. We therefore choose a low-DOF BSpline model. | ||
| + | *Because the implementation of the BRAINSfit module used here expects a 3D volume, we choose a slightly larger grid and make it isotropic, although the 3rd dimension of the grid is not used. We expect the optimizer in the registration to not move outside the slice plane and simply return 0 deformations there. | ||
| + | *An alternative strategy would be to register each image to its immediate previous timepoint, and then integrating the displacements. This has the advantage that nonrigid registration tends to be better for smaller differences, but it bears the risk of accumulating error and hence a lower robustness. | ||
| + | *A related option to improve precision would be to initialize each registration with the result of the previous one. Unfortunately the BRAINSfit registration modules do not (yet) allow to initialize with anything other than an affine. | ||
| + | *The additional information/constraint, that the displacements defined by the series of registrations should describe a smooth motion that is contiguous and differentiable, has not been exploited in this approach. This could be done a posteriori by applying a temporal filter across the transforms or fitting an appropriate model curve to each node displacement. | ||
| + | *Overall strategy: | ||
| + | :#select one frame within the breathing cycle as reference frame | ||
| + | :#compute non-rigid BSpline registration of all images to the reference frame | ||
| + | :#extract the displacements of indiv. grid nodes and plot over time | ||
| + | === Discussion: Registration Challenges === | ||
| + | *large differences in FOV | ||
| + | *strong differences in image contrast between MRI & CT | ||
| + | *contrast enhancement and pathology and treatment changes cause additional differences in image content | ||
| + | *we have strongly anisotropic voxel sizes with much less through-plane resolution | ||
| + | |||
| + | |||
| + | === Procedures === | ||
| + | *'''Phase I: Pilot to determine optimal registration parameters''' | ||
| + | #load reference image and one moving image from the series | ||
| + | #open Registration : ''BrainsFit'' module | ||
| + | ##Registration Phases: | ||
| + | ##set "reference" fixed and "moving_??" as moving image | ||
| + | ###select/check ''Include BSpline registration phase'' | ||
| + | ##Output Settings: | ||
| + | ###select a new transform "Slicer BSpline Transform", rename to "Xf1_moving_??" | ||
| + | ###select a new volume "Output Image Volume'', rename to "moving_??_Xf1" | ||
| + | ##Registration Parameters: increase ''Number Of Samples'' to 200,000 | ||
| + | ##Registration Parameters: set ''Number Of Grid Subdivisions'' to 5,5,5 | ||
| + | ##Leave all other settings at default | ||
| + | ##click: Apply; (runtime < 10 sec. on MacPro) | ||
| + | ##adjust grid size until registration is acceptable | ||
| + | ##you can see the commandline text of the registration performed by opening the ''Window/Error Log'' window and clicking on ''BRAINSfit commandline input'' | ||
| + | *'''Phase II: Batch Run''' | ||
| + | #open a terminal window | ||
| + | ##via a TextEditor or prototyping/scripting software (e.g. Matlab), copy and modify the prototype line below, by changing only the moving input image: | ||
| + | /Applications/Slicer36/Slicer3 --launch /Applications/Slicer36/lib/Slicer3/Plugins/BRAINSFit --useBSpline --splineGridSize 5,5,5 --outputVolumePixelType short / | ||
| + | --numberOfSamples 200000 --costMetric MMI --fixedVolume Reference/refLung_001.dcm --movingVolume Moving/Moving_001/Moving_001.dcm / | ||
| + | --bsplineTransform Xforms/Moving_001_XfBSpl5.tfm --outputVolume MovingResampled/Moving_001_r.nrrd >> Logs/Moving_001_RegLog.txt 2>&1 | ||
| + | ##'''replace "/Applications/Slicer36" with your path of 3DSlicer''' | ||
| + | ## create result directories '''MovingResampled''', '''Logs''', '''Xforms''' | ||
| + | ## note that because input image is DICOM, and images are 2D only, each image of the time series must be in its own directory, otherwise Slicer will read them as a volume. | ||
| + | ##paste all commands into a terminal window, or copy into a shell script and execute. | ||
| + | *'''Phase III: Evaluate Transform files''' | ||
| + | ##upon completion, read the transform files with an editor and extract the displacements of interest | ||
| + | ##The ITK transform files describe displacements at the grid nodes, many of which are outside the region of interest. Because BRAINSfit pads the grid with 1 voxel, the grid returned is actually 8x8x8. We use only the plane (*,*,4) for analysis and discard the y-direction displacements, which as expected are all zero. | ||
| + | ##for details on the ITK transform format see the [http://www.slicer.org/slicerWiki/index.php/Slicer-3-6-FAQ#What_is_the_Meaning_of_.27Fixed_Parameters.27_in_the_transform_file_.28.tfm.29_of_a_BSpline_registration_.3F '''FAQ''' here] | ||
| + | ##To obtain displacements at arbitrary coordinates, interpolate the transform into a deformation field, e.g. using this module (details [http://www.slicer.org/slicerWiki/index.php/Slicer-3-6-FAQ#How_can_I_convert_a_BSpline_transform_into_a_deformation_field.3F FAQ here]): | ||
| + | /Applications/Slicer3.6.3/lib/Slicer3/Plugins/BSplineToDeformationField | ||
| − | |||
| − | |||
=== Registration Results=== | === Registration Results=== | ||
| − | [ | + | {| style="color:#bbbbbb; " cellpadding="10" cellspacing="0" border="0" |
| + | |[[Image:RegLib_C46_Thumb2.gif|300px|lleft|moving input]] | ||
| + | |[[Image:RegLib_C46_Moving_animReg.gif|300px|lleft|moving input after registration (only frames 1-27 shown)]] | ||
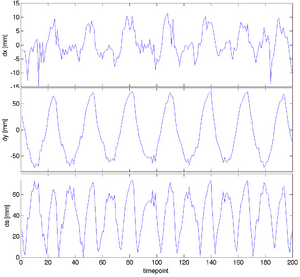
| + | |[[Image:RegLib_C46_Displacements.png|300px|lleft|displacements]] | ||
| + | |- | ||
| + | |unregistered moving series | ||
| + | |moving input after registration (only frames 1-27 shown) | ||
| + | |[[Media:RegLib_C46_Displacements.pdf|displacements (PDF)]] | ||
| + | |} | ||
| + | |||
| + | |||
| + | |||
===Download === | ===Download === | ||
| + | *Data | ||
*[[Media:RegLib_C12_LiverTumor_DATA.zip|'''download input image data''' <small> (Input Data, NRRD images, zip file 42 MB) </small>]] | *[[Media:RegLib_C12_LiverTumor_DATA.zip|'''download input image data''' <small> (Input Data, NRRD images, zip file 42 MB) </small>]] | ||
*[[Media:RegLib_C12_LiverTumorAblation1_ParameterPresets.mrml|'''download registration parameter presets file''' <small> (.mrml file 20 kB) </small>]] | *[[Media:RegLib_C12_LiverTumorAblation1_ParameterPresets.mrml|'''download registration parameter presets file''' <small> (.mrml file 20 kB) </small>]] | ||
*[[Media:RegLib_C12_LiverTumorAblation1_Tutorial.ppt|'''download guided tutorial''' <small> (PowerPoint, xx MB) </small>]] | *[[Media:RegLib_C12_LiverTumorAblation1_Tutorial.ppt|'''download guided tutorial''' <small> (PowerPoint, xx MB) </small>]] | ||
*[[Media:RegLib_C12_LiverTumorAblation1_Set.zip|'''download full tutorial set''' <small> (Input Data, presets, results, tutorial, zip file xx MB) </small>]] | *[[Media:RegLib_C12_LiverTumorAblation1_Set.zip|'''download full tutorial set''' <small> (Input Data, presets, results, tutorial, zip file xx MB) </small>]] | ||
| − | |||
[[Projects:RegistrationDocumentation:ParameterPresetsTutorial|Link to User Guide: How to Load/Save Registration Parameter Presets]] | [[Projects:RegistrationDocumentation:ParameterPresetsTutorial|Link to User Guide: How to Load/Save Registration Parameter Presets]] | ||
| − | < | + | *Results |
| − | + | **[[Media:RegLib_C46_RegistrationTransforms.zip |'''RegLib_C46_RegistrationTransforms.zip''' : the 200 BSpline registration transforms <small> (ITK transform (text) files, zip file 274 kB) </small>]]''' | |
| − | + | **[[Media:RegLib_C46_MovingResampled.zip |'''RegLib_C46_MovingResampled.zip''' : the 200 registered image stack <small> (NRRD image files, zip file 16MB) </small>]]''' | |
| − | |||
| − | |||
| − | * | ||
| − | * | ||
| − | |||
| − | |||
| − | === | + | === Acknowledgments === |
| − | + | Thanks to Dr.Stuart Silverman and Dr. Nobuhiko Hata for sharing this case. | |
Revision as of 15:56, 18 August 2011
Home < Projects:RegistrationLibrary:RegLib C12Back to ARRA main page
Back to Registration main page
Back to Registration Use-case Inventory
Contents
v3.6.3  Slicer Registration Library Case #12: Liver Tumor Cryoablation
Slicer Registration Library Case #12: Liver Tumor Cryoablation
Input

|
|
|

|
|
| fixed image/target | intermediate ref. image | moving image |
Modules
- Slicer 3.6.3 recommended modules: BrainsFit
Objective / Background
We seek to align a pre-operative MRI with the intra-operative CT to enhance tumor visibility for surgical guidance.
Keywords
MRI, CT, IGT, intra-operative, liver, cryoablation, change detection, non-rigid registration
Input Data
- reference/fixed : pr-op CT, 0.95 x 0.95 x 5 mm voxel size
- moving: intra-op MRI, 0.78 x 0.78 x 2.5 mm axial,
Notes / Overall Strategy
- because the torso is already well registered and does not move across the sequence, a global affine/rigid registration is of no use. Instead we seek to assess the internal displacements caused across the breathing cycle. We therefore choose a low-DOF BSpline model.
- Because the implementation of the BRAINSfit module used here expects a 3D volume, we choose a slightly larger grid and make it isotropic, although the 3rd dimension of the grid is not used. We expect the optimizer in the registration to not move outside the slice plane and simply return 0 deformations there.
- An alternative strategy would be to register each image to its immediate previous timepoint, and then integrating the displacements. This has the advantage that nonrigid registration tends to be better for smaller differences, but it bears the risk of accumulating error and hence a lower robustness.
- A related option to improve precision would be to initialize each registration with the result of the previous one. Unfortunately the BRAINSfit registration modules do not (yet) allow to initialize with anything other than an affine.
- The additional information/constraint, that the displacements defined by the series of registrations should describe a smooth motion that is contiguous and differentiable, has not been exploited in this approach. This could be done a posteriori by applying a temporal filter across the transforms or fitting an appropriate model curve to each node displacement.
- Overall strategy:
- select one frame within the breathing cycle as reference frame
- compute non-rigid BSpline registration of all images to the reference frame
- extract the displacements of indiv. grid nodes and plot over time
Discussion: Registration Challenges
- large differences in FOV
- strong differences in image contrast between MRI & CT
- contrast enhancement and pathology and treatment changes cause additional differences in image content
- we have strongly anisotropic voxel sizes with much less through-plane resolution
Procedures
- Phase I: Pilot to determine optimal registration parameters
- load reference image and one moving image from the series
- open Registration : BrainsFit module
- Registration Phases:
- set "reference" fixed and "moving_??" as moving image
- select/check Include BSpline registration phase
- Output Settings:
- select a new transform "Slicer BSpline Transform", rename to "Xf1_moving_??"
- select a new volume "Output Image Volume, rename to "moving_??_Xf1"
- Registration Parameters: increase Number Of Samples to 200,000
- Registration Parameters: set Number Of Grid Subdivisions to 5,5,5
- Leave all other settings at default
- click: Apply; (runtime < 10 sec. on MacPro)
- adjust grid size until registration is acceptable
- you can see the commandline text of the registration performed by opening the Window/Error Log window and clicking on BRAINSfit commandline input
- Phase II: Batch Run
- open a terminal window
- via a TextEditor or prototyping/scripting software (e.g. Matlab), copy and modify the prototype line below, by changing only the moving input image:
/Applications/Slicer36/Slicer3 --launch /Applications/Slicer36/lib/Slicer3/Plugins/BRAINSFit --useBSpline --splineGridSize 5,5,5 --outputVolumePixelType short / --numberOfSamples 200000 --costMetric MMI --fixedVolume Reference/refLung_001.dcm --movingVolume Moving/Moving_001/Moving_001.dcm / --bsplineTransform Xforms/Moving_001_XfBSpl5.tfm --outputVolume MovingResampled/Moving_001_r.nrrd >> Logs/Moving_001_RegLog.txt 2>&1
- replace "/Applications/Slicer36" with your path of 3DSlicer
- create result directories MovingResampled, Logs, Xforms
- note that because input image is DICOM, and images are 2D only, each image of the time series must be in its own directory, otherwise Slicer will read them as a volume.
- paste all commands into a terminal window, or copy into a shell script and execute.
- Phase III: Evaluate Transform files
- upon completion, read the transform files with an editor and extract the displacements of interest
- The ITK transform files describe displacements at the grid nodes, many of which are outside the region of interest. Because BRAINSfit pads the grid with 1 voxel, the grid returned is actually 8x8x8. We use only the plane (*,*,4) for analysis and discard the y-direction displacements, which as expected are all zero.
- for details on the ITK transform format see the FAQ here
- To obtain displacements at arbitrary coordinates, interpolate the transform into a deformation field, e.g. using this module (details FAQ here):
/Applications/Slicer3.6.3/lib/Slicer3/Plugins/BSplineToDeformationField
Registration Results
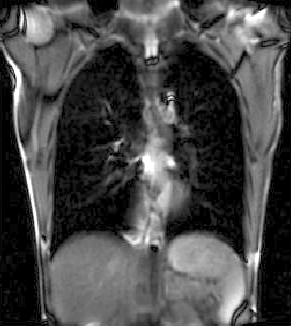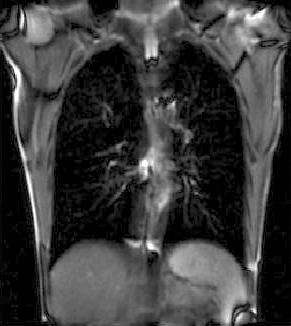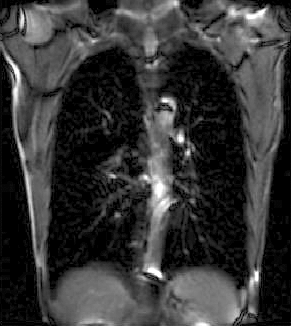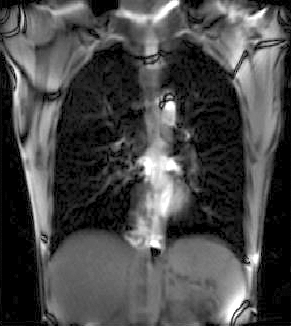
|
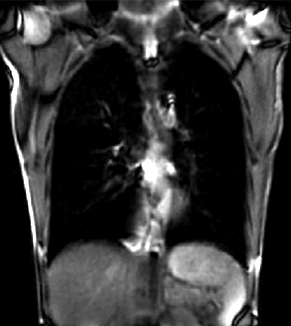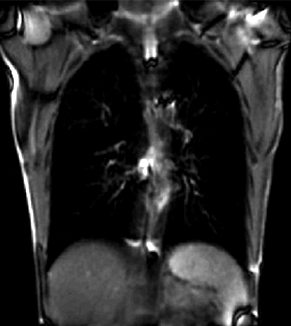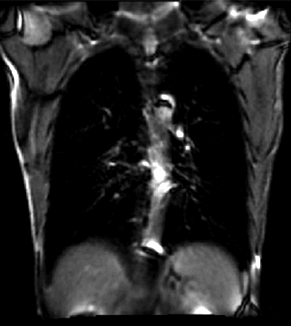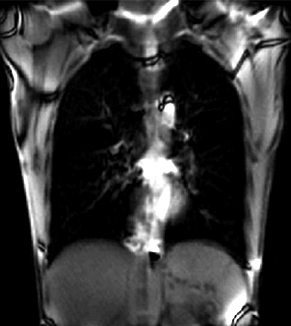
|
|
| unregistered moving series | moving input after registration (only frames 1-27 shown) | displacements (PDF) |
Download
- Data
- download input image data (Input Data, NRRD images, zip file 42 MB)
- download registration parameter presets file (.mrml file 20 kB)
- download guided tutorial (PowerPoint, xx MB)
- download full tutorial set (Input Data, presets, results, tutorial, zip file xx MB)
Link to User Guide: How to Load/Save Registration Parameter Presets
- Results
Acknowledgments
Thanks to Dr.Stuart Silverman and Dr. Nobuhiko Hata for sharing this case.