Difference between revisions of "Projects:RegistrationLibrary:RegLib C01b"
From NAMIC Wiki
m (Text replacement - "http://www.slicer.org/slicerWiki/index.php/" to "https://www.slicer.org/wiki/") |
|||
| Line 20: | Line 20: | ||
=== Slicer 4.1 recommended Modules === | === Slicer 4.1 recommended Modules === | ||
| − | *[ | + | *[https://www.slicer.org/wiki/Modules:BRAINSFit BrainsFit] |
| − | *[ | + | *[https://www.slicer.org/wiki/Modules:AffineRegistration-Documentation-3.6 Fast Affine Registration] |
| − | *[ | + | *[https://www.slicer.org/wiki/Modules:RegisterImages-Documentation-3.6 Expert Automated Registration] |
===Objective / Background === | ===Objective / Background === | ||
Latest revision as of 17:36, 10 July 2017
Home < Projects:RegistrationLibrary:RegLib C01bBack to ARRA main page
Back to Registration main page
Back to Registration Case Library
v3.6.3  Slicer Registration Library Case #01:
Slicer Registration Library Case #01:
Intra-subject Brain MRI: axial T1 Tumor Growth Assessment
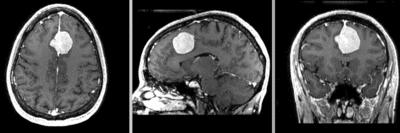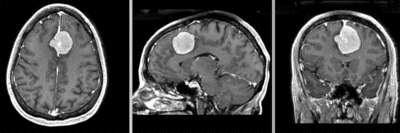
Input
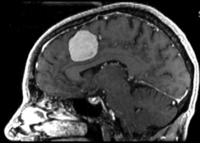
|
|
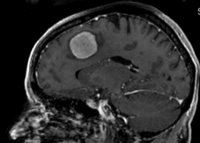
|
| fixed image/target | moving image |
Versions
For the Slicer 4.1 version of this tutorial see here
Slicer 4.1 recommended Modules
Objective / Background
This is a classic case of change assessment. We want to know if the tumor changed since last exam.
Keywords
MRI, brain, head, intra-subject, T1, tumor growth, meningioma, change assessment
Input Data
- reference/fixed : T1 SPGR , 0.9375 x 0.9375 x 1.4 mm voxel size, axial, RAS orientation.
- moving: T1 SPGR , 0.9375 x 0.9375 x 1.2 mm voxel size, sagittal, RAS orientation.
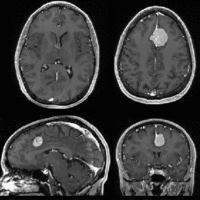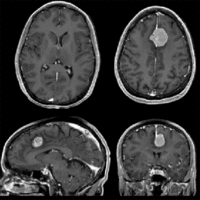
Registration Results
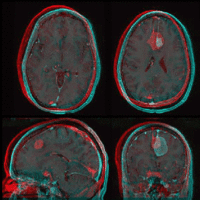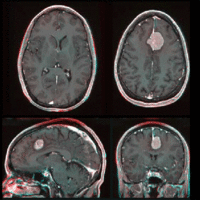
| Fast Affine registration result | ExpertAutomatedRegistration registration result | before/after registration with ExpertAutomatedRegistration module |
Download
- Slicer 3.6.3 packages:
- Slicer 3.4 packages:
- download entire package (Data,Presets,Tutorial, Solution, zip file 33.7 MB)
- download registration parameter presets file (MRML file, import as scene)
- download image dataset only (NRRD, 10.7 MB, filename: RegLib_C01_Data_TumorGrowth.zip)
- download image dataset in NIFTI format (NIFTI / nii, 10.7 MB, filename: RegLib_C01_DataNIFTI_TumorGrowth.zip)
- result transform file (ITK .tfm file, load into slicer and apply to the target volume)
- Tutorials (step-by -step walk through):
- Multiresolution testresult package (Data,Xform, Solution, zip file 16.5 MB)
- Download package from MIDAS server (Data,Xform)
Link to User Guide: How to Load/Save Registration Parameter Presets
Discussion: Registration Challenges
- accuracy is the critical criterion here. We need the registration error (residual misalignment) to be smaller than the change we want to measure/detect. Agreement on what constitutes good alignment can therefore vary greatly.
- the two images have strong differences in coil inhomogeneity. This affects less the registration quality but hampers evaluation. Most of the difference does not become apparent until after registration in direct juxtaposition. Bias field correction beforehand is recommended.
- we have slightly different voxel sizes
- if the pathology change is substantial it might affect the registration.
Discussion: Key Strategies
- the two images have identical contrast, hence we consider "sharper" cost functions, such as NormCorr or MeanSqrd
- general practice is to register the follow-up to the baseline. However here the follow-up has slightly higher resolution. From an image quality/data perspective it would be better to use the highest resolution image as your fixed/reference. But here follow the most common convention, i.e. fixed image is the baseline.
- because we seek to assess/quantify regional size change, we must use a rigid (6DOF) scheme, i.e. we must exclude scaling.
- if the pathology change is soo large that it might affect the registration, we should mask it out. The simplest way to do this is to build a box ROI from the ROItool and feed it as input to the registration. Remember that masking does not mean that masked areas aren't matched, they just do not contribute to the cost function driving the registration, but move along passively. Next more involved level would be to outline the tumor. If a segmentation is available, you can use that.
- these two images are not too far apart initially, so we reduce the default of expected translational misalignment
- because accuracy is more important than speed here, we increase the sampling rate from the default 2% to 15%.
- we also expect minimal differences in scale & distortion: so we can either set the expected values to 0 or run a rigid registration
- we test the result in areas with good anatomical detail and contrast, far away from the pathology. With rigid body motion a local measure of registration accuracy is representative and can give us a valid limit of detectable change.