Difference between revisions of "Algorithm:Utah"
| Line 41: | Line 41: | ||
|- | |- | ||
| − | | | | + | | | [[Image:brain-seg-utah.tif|200px]] |
| | | | | | ||
Revision as of 19:50, 1 January 2008
Home < Algorithm:UtahBack to NA-MIC Algorithms
Overview of Utah Algorithms
We are developing new methods in the areas of statistical shape analysis, MRI tissue segmentation, and diffusion tensor image processing and analysis. We are building shape analysis tools that can generate efficient statistical models appropriate for analyzing anatomical shape differences in the brain. We are developing a wide range of tools for diffusion tensor imaging, that span the entire pipeline from image processing to automatic white matter tract extraction to statistical testing of clinical hypotheses.
Utah Projects
|
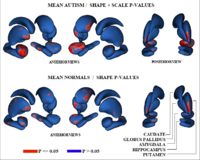
Adaptive, Particle-Based Sampling for Shapes and ComplexesThis research is a new method for constructing compact statistical point-based models of ensembles of similar shapes that does not rely on any specific surface parameterization. The method requires very little preprocessing or parameter tuning, and is applicable to a wider range of problems than existing methods, including nonmanifold surfaces and objects of arbitrary topology. More... New: J Cates, PT Fletcher, M Styner, M Shenton, R Whitaker, Shape modeling and analysis with entropy-based particle systems, IPMI 2007, pp. 333-345. |
|
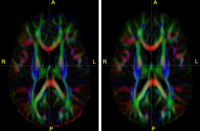
Diffusion Tensor Image Processing ToolsWe implement the diffusion weighted image (DWI) registration model from the paper of G.K.Rohde et al. Patient head motion and eddy currents distortion cause artifacts in maps of diffusion parameters computer from DWI. This model corrects these two distortions at the same time including brightness correction. New: We have recently developed software for eddy current correction. |
|
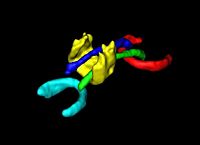
DTI Volumetric White Matter ConnectivityWe have developed a PDE-based approach to white matter connectivity from DTI that is founded on the principal of minimal paths through the tensor volume. Our method computes a volumetric representation of a white matter tract given two endpoint regions. We have also developed statistical methods for quantifying the full tensor data along these pathways, which should be useful in clinical studies using DT-MRI. More... New: PT Fletcher, R Tao, W-K Jeong, RT Whitaker, A volumetric approach to quantifying region-to-region white matter connectivity in diffusion tensor MRI, IPMI 2007, pp. 346-358. |
| File:Brain-seg-utah.tif |
Tissue Classification with Neighborhood StatisticsWe have implemented an MRI tissue classification algorithm based on unsupervised non-parametric density estimation of tissue intensity classes. More... T Tasdizen, S Awate, R Whitaker, A nonparametric, entropy-minimizing MRI tissue classification algorithm implementation using ITK, MICCAI 2005 Open-Source Workshop. |