Difference between revisions of "Projects:MultimodalAtlas"
| Line 1: | Line 1: | ||
Back to [[NA-MIC_Internal_Collaborations:StructuralImageAnalysis|NA-MIC Collaborations]], [[Algorithm:MIT|MIT Algorithms]], [[DBP2:Harvard|Harvard DBP2]] | Back to [[NA-MIC_Internal_Collaborations:StructuralImageAnalysis|NA-MIC Collaborations]], [[Algorithm:MIT|MIT Algorithms]], [[DBP2:Harvard|Harvard DBP2]] | ||
__NOTOC__ | __NOTOC__ | ||
| − | + | Today, computational anatomy studies are mainly hypothesis-driven, aiming to identify and characterize structural or functional differences between, for instance a group of patients with a specific disorder and control subjects. This type of approach has two premises: clinical classification of the subjects and spatial correspondence across the images. In practice, achieving either can be challenging. First, the complex spectrum of symptoms of neuro-degenerative disorders like schizophrenia and overlapping symptoms across different types of dementia like Alzheimer's disease, delirium and depression make a diagnosis based on standardized clinical tests like the mental status examination difficult. Second, across-subject correspondence in the images is a particularly hard problem that requires different approaches in various contexts. A popular technique is to normalize all subjects into a standard space, such as the Talairach space, by registering each image with a single, universal template image that represents an average brain. However, the quality of such an approach is limited by the accuracy with which the universal template represents the population in the study. | |
| − | + | With the increasing availability of medical images, data-driven algorithms offer the ability to probe a population and potentially discover sub-groups that may differ in unexpected ways. In this paper, we propose and demonstrate an efficient probabilistic clustering algorithm, called '''iCluster''', that: | |
| + | |||
| + | * computes a small number of templates that summarize a given population of images, | ||
| + | * simultaneously co-registers all the images using a nonlinear transformation model, | ||
| + | * assigns each input image to a template. | ||
| + | |||
| + | The templates are guaranteed to live in an affine-normalized space, i.e., they are spatially aligned with respect to an affine transformation model. | ||
= Description = | = Description = | ||
Revision as of 20:09, 16 May 2008
Home < Projects:MultimodalAtlasBack to NA-MIC Collaborations, MIT Algorithms, Harvard DBP2
Today, computational anatomy studies are mainly hypothesis-driven, aiming to identify and characterize structural or functional differences between, for instance a group of patients with a specific disorder and control subjects. This type of approach has two premises: clinical classification of the subjects and spatial correspondence across the images. In practice, achieving either can be challenging. First, the complex spectrum of symptoms of neuro-degenerative disorders like schizophrenia and overlapping symptoms across different types of dementia like Alzheimer's disease, delirium and depression make a diagnosis based on standardized clinical tests like the mental status examination difficult. Second, across-subject correspondence in the images is a particularly hard problem that requires different approaches in various contexts. A popular technique is to normalize all subjects into a standard space, such as the Talairach space, by registering each image with a single, universal template image that represents an average brain. However, the quality of such an approach is limited by the accuracy with which the universal template represents the population in the study.
With the increasing availability of medical images, data-driven algorithms offer the ability to probe a population and potentially discover sub-groups that may differ in unexpected ways. In this paper, we propose and demonstrate an efficient probabilistic clustering algorithm, called iCluster, that:
- computes a small number of templates that summarize a given population of images,
- simultaneously co-registers all the images using a nonlinear transformation model,
- assigns each input image to a template.
The templates are guaranteed to live in an affine-normalized space, i.e., they are spatially aligned with respect to an affine transformation model.
Description
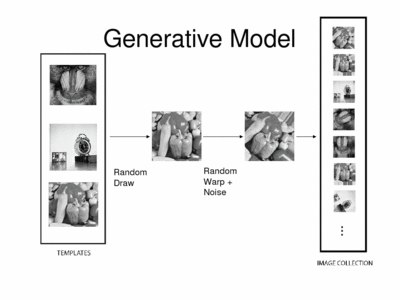
iCluster is derived from a simple generative model. We assume that there are a fixed and known number of template images. Then the process that generates an observed image is as follows: a template is randomly drawn – note that the probability that governs this process doesn’t have to be uniform. Next, the chosen template is warped with a random transformation and i.i.d Gaussian noise is added to this warped image to generate an observed image. This process is repeated multiple times to generate a collection of images.
We formulate the problem as a maximum likelihood solution. We employ a Generalized Maximum Likelihood algorithm to solve the problem.
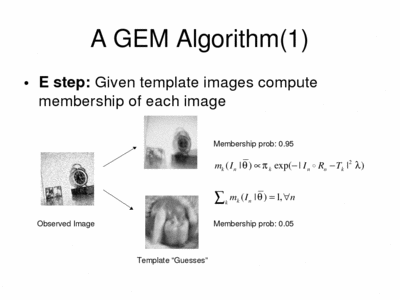
In the E-step, the algorithm computes membership probabilities.
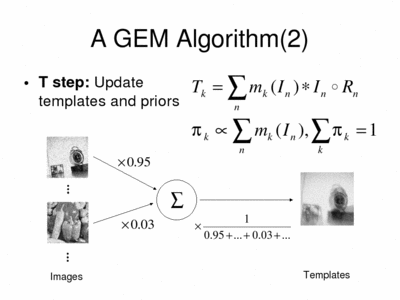
We divide the M-step into two sub-steps: the first one, called the T (template) step is where we have closed-form expressions for the optimization. The algorithm updates the template images as weighted averages of the images, where the weights are the memberships. The template priors are updated as normalized sums of memberships.
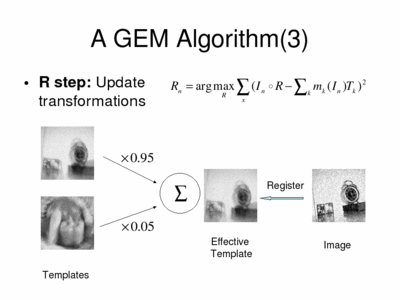
In the second part of the M-step, called R (registration) step, the algorithm performs a collection of independent pairwise registration instances. Each image is registered with a unique, "effective" template, which is a weighted average of the current templates and the weights are the corresponding memberships.
Results
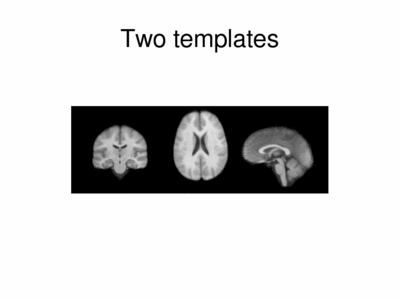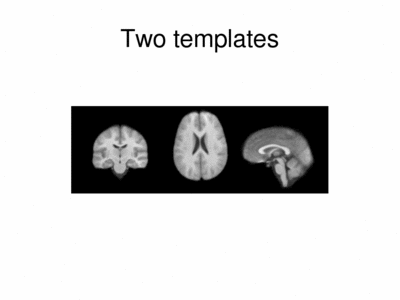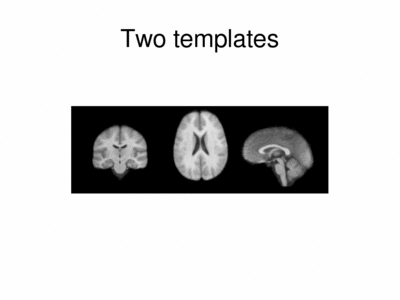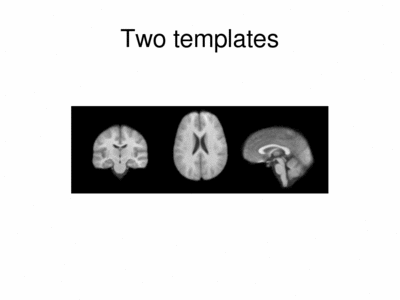
We ran iCluster with k = 2 on a 50 subject MR data set. The following shows the two templates (modes) discovered by the algorithm.
'Software'
The algorithm is currently implemented in the Insight ToolKit (ITK) and will be made publicly available. We also plan to integrate it into Slicer.
Key Investigators
MIT Algorithms: Mert R. Sabuncu and Polina Golland
Harvard DBP 2: M.E. Shenton, M. Kubicki and S. Bouix
Publications
In Print