Difference between revisions of "DBP2:Harvard:Brain Segmentation Roadmap"
(→Module) |
|||
| Line 66: | Line 66: | ||
:* Module documentation can be found here: | :* Module documentation can be found here: | ||
:**[[Media:IJdata.tar.gz|Training Dataset]] | :**[[Media:IJdata.tar.gz|Training Dataset]] | ||
| − | :**[[Media: | + | :**[[Media:PNLStochasictNew.ppt|Training Presentation]] |
:**[[Media:Helix.zip|Sample Helix Dataset]] | :**[[Media:Helix.zip|Sample Helix Dataset]] | ||
:* Software that you will also need to launch: | :* Software that you will also need to launch: | ||
| Line 73: | Line 73: | ||
:**[http://pnl.bwh.harvard.edu/NAMIC/Slicer3-build.zip Slicer 3 Build for Windows] | :**[http://pnl.bwh.harvard.edu/NAMIC/Slicer3-build.zip Slicer 3 Build for Windows] | ||
:**[http://pnl.bwh.harvard.edu/NAMIC/Slicer3-lib.zip Slicer 3 Lib for Windows] | :**[http://pnl.bwh.harvard.edu/NAMIC/Slicer3-lib.zip Slicer 3 Lib for Windows] | ||
| + | |||
| + | |||
| + | |||
| + | |||
==Work Accomplished== | ==Work Accomplished== | ||
Revision as of 22:36, 7 January 2009
Home < DBP2:Harvard:Brain Segmentation RoadmapBack to NA-MIC Collaborations, Harvard DBP 2
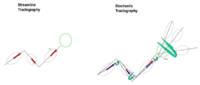
Stochastic Tractography for VCFS
Roadmap
The main goal of this project is to develop end-to-end application that would be used to characterize anatomical connectivity abnormalities in the brain of patients with velocardiofacial syndrome (VCFS), and to link this information with deficits in schizophrenia. This page describes the technology roadmap for stochastic tractography, using newly acquired 3T data, NAMIC tools and slicer 3.
Algorythm
|
|
- C-References
- Björnemo M, Brun A, Kikinis R, Westin CF. Regularized stochastic white matter tractography using diffusion tensor MRI. In Fifth International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI'02). Tokyo, Japan, 2002;435-442.
- Friman, O., Farneback, G., Westin CF. A Bayesian Approach for Stochastic White Matter Tractography. IEEE Transactions on Medical Imaging, Vol 25, No. 8, Aug. 2006
Module
Can be found in: MODULES > PYTHON MODULES > PYTHON STOCHASTIC TRACTOGRAPHY Functionality of Python Stochastic Tractography module in Slicer 3.0
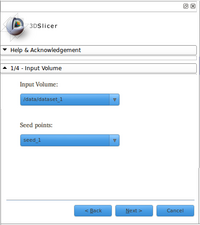
- Loading volumes:
Module reads files (DWI and ROIs) in nhdr format.
- Smoothing:
One can smooth the DWI data (only Gausian smoothing is supported at this time). We recommend it if the data is noisy.
- Masks:
Masks are removing some of the unwanted tracts, same way principal diffusion direction tractography uses stopping criteria for tractography termination. Three different methods for masking are provided.
- 1. Otsu Mask- a level-set method creating brain mask.
- 2. WM Mask- a thresholding method based on B0 intensity.
- 3. Artifact removal- one can use its own WM mask, obtained from T1 or T2 segmentation, and registered to DWI space, then artifact removal compares WM mask with FA map, and patches WM mask with voxels that have high FA values.
- Tensor estimation:
Module uses two methods for estimating tensors, and can output anisotropy indices (FA/Mode/Trace), if requested
- Stochastic Tractography:
Parameters that need to be adjusted:
- 1. The amount of tracts that will be seeded from each voxel (we recommend between 500 and 1000 tracts, depending on the workstation power- 1000 tracts per voxel seeded within the large ROI for high resolution DWI can take a long time to compute).
- 2. Maximum tract length (in mm), this can eliminate long, unwanted tracts if the regions for which connection is measured are located close to each other
- 3. Step Size: distance between each re-estimation of tensors, usually between 0.5 and 1 mm
- 4. Stopping criteria. This can be used on the top of WM mask, to terminate tracts (in case they really want to travel through the CSF, for example).
- Probability Map:
This step creates output probability maps.
- 1. rough: each voxel is counted only once if at least one fiber pass through it
- 2. cumulative:Tracts are summed by voxel independently
- 3. discriminative: tracts are summed by voxel depending on their length
Then, probability maps can be saved as ROIs, and either used directly, or thresholded (at certain probability, step claimed by few publications to remove noise) in slicer to mask and compute average FA, Mode, Trace for entire connection. Diffusion indices can be also weighted by the probability of connection for each voxel.
- Module documentation can be found here:
- Software that you will also need to launch:
Work Accomplished
- A - Optimization and testing of stochastic tractography algorythm
-
- Original methodological paper, as well as our first attempts to use the algorythm (CC+ and matlab scripts) have been done on old "NAMIC" 1.5T LSDI data (Structural MRI and DTI data).
- Tri worked hard on making sure algorythm works on new high resolution 3T data (available here: 3T Data).
- Tests have been done also on the spiral diffusion phantom, to make sure diffusion directions and scanner coordinates are handled properly by the algorythm (Figure 6).
Work in Progress
|
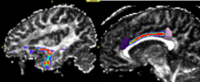
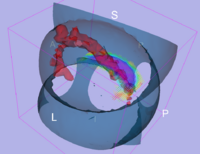
We have started the project of investigating Arcuate Fasciculus using Stochastic Tractography (Figure 7). This structure is especially important in both VCFS and schizophrenia, as it connects language related areas (Brocka and Wernicke's), and is involved in language processing quite disturbed in schizophrenia patients. It also can not be reliably traced using deterministic tractography.
|
We use combination of fMRI and DTI data to define and characterize functional and anatomical connectivity within the semantic processing network in schizophrenia.
We have started collaboration with Department of Neurophysiology Max Planck Institute in Frankfurt (Anna Rotarska contact person). They have dataset containing DTI and resting state fMRI in schizophrenia, and want to use stochastic tractography to measure integrity of anatomical connections within the default network in schizophrenia. Their fMRI data has been co-registered with anatomical scans, and we are putting it into the DTI space. Once this is done, we will start creating tracts connecting fMRI ROIs. This data will be also used to test robustness of our module, since data was collected on a different scanner (3T Philips).
We are also working on a tractography comparison projectdataset, where we apply stochastic tractography to phantom, as well as test dataset. Staffing Plan
|
Schedule
- 10/2007 - Optimization of Stochastic Tractography algorythm for 1.5T data.
- 10/2007 - Algorythm testing on Santa Fe data set and diffusion phantom.
- 06/2008 - Optimization of Stochastic Tractography algorythm for 3T data.
- 11/2008 - Slicer 3 module prototype using python.
- 12/2008 - Slicer 3 module official release
- 12/2008 - Documentation and packaging for dissemination.
- 12/2008 - Arcuate Fasciculus results.
- 01/2009 - Arcuate Fasciuclus first draft of the paper.
- 05/2009 - Distortion correction and nonlinear registration added to the module
- 05/2009 - Symposium on tractography, including stochastic methods at World Biological Psychiatry Symposium in Florence, Italy.
- 05/2009 - Presentation of Arcuate Fasciculus findings at World Biological Psychiatry Symposium in Florence, Italy.
Team and Institute
- PI: Marek Kubicki (kubicki at bwh.harvard.edu)
- DBP2 Investigators: Sylvain Bouix, Yogesh Rathi, Julien de Siebenthal
- NA-MIC Engineering Contact: Brad Davis, Kitware
- NA-MIC Algorithms Contact: Polina Gollard, MIT
Publications
In print