Difference between revisions of "2010 Winter Project Week Musco Skeletal Segmentation"
| Line 29: | Line 29: | ||
<i><u>Approach:</u></i> | <i><u>Approach:</u></i> | ||
<BR> | <BR> | ||
| − | Multi-Contrast MR images are collected and seed points for each region of interest are taken as input. Cluster center and standard deviation are | + | Multi-Contrast MR images are collected and seed points for each region of interest are taken as input. Cluster center and standard deviation are calculated for each ROI based on pixel intensities of the seed points. The pixels are clustered based on different pixel intensity values in multiple MR images to the nearest cluster center radius. |
For detailed approach, please visit http://www.na-mic.org/Wiki/index.php/Stanford_Simbios_group | For detailed approach, please visit http://www.na-mic.org/Wiki/index.php/Stanford_Simbios_group | ||
| Line 46: | Line 46: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| + | Finished segmenting different regions of interest like bones, cartilage etc. | ||
| + | Created label maps from existing segmented output. | ||
</div> | </div> | ||
</div> | </div> | ||
Revision as of 20:24, 23 December 2009
Home < 2010 Winter Project Week Musco Skeletal Segmentation
Key Investigators
- Stanford: Harish Doddi, Saikat Pal, Scott Delp
- Kitware: Luis Ibanez
Objective
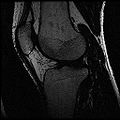
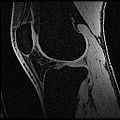
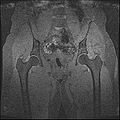
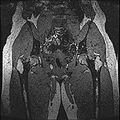
The aim of this project is to develop an semi-automatic rapid methodology to convert whole body imaging datasets into three-dimensional geometries for modeling simulations. We have used a multi-contrast MR Segmentation approach to cluster the structures of interest. The output label maps will be smoothed and existing surface meshes will be morphed on to a target image.
Approach, Plan
Approach:
Multi-Contrast MR images are collected and seed points for each region of interest are taken as input. Cluster center and standard deviation are calculated for each ROI based on pixel intensities of the seed points. The pixels are clustered based on different pixel intensity values in multiple MR images to the nearest cluster center radius.
For detailed approach, please visit http://www.na-mic.org/Wiki/index.php/Stanford_Simbios_group
Plan:
a. Smooth the existing segmented label maps
b. Build models for bones and cartilage from the existing segmented label maps.
Progress
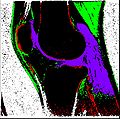
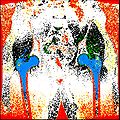
Finished segmenting different regions of interest like bones, cartilage etc.
Created label maps from existing segmented output.