Difference between revisions of "Projects:RegistrationLibrary:RegLib C02"
| Line 85: | Line 85: | ||
[[Image:RegLib_C02_Result_AnimGif.gif|500px|Robust Multiresolution Registration Result: FLAIR + segmentation aligned with SPGR]] : registered w. Multiresolution affine <br> | [[Image:RegLib_C02_Result_AnimGif.gif|500px|Robust Multiresolution Registration Result: FLAIR + segmentation aligned with SPGR]] : registered w. Multiresolution affine <br> | ||
[[Image:RegLib_C02_AGif_BrainsFit.gif|500px|BrainsFit Result: FLAIR aligned with SPGR]]: registered w. BrainsFit <br> | [[Image:RegLib_C02_AGif_BrainsFit.gif|500px|BrainsFit Result: FLAIR aligned with SPGR]]: registered w. BrainsFit <br> | ||
| + | <br> | ||
| + | '''Conclusion:''' Both Multiresolution and BrainsFit modules produce good registrations without the need for additional initialization or masking. The BrainsFit solution is the faster of the two (3 minutes vs. 15 minutes). | ||
=== Download === | === Download === | ||
Revision as of 17:14, 25 August 2010
Home < Projects:RegistrationLibrary:RegLib C02Back to ARRA main page
Back to Registration main page
Back to Registration Use-case Inventory
Contents
v3.6.1  Slicer Registration Library Case #02: Intra-subject Brain MR FLAIR to MR T1
Slicer Registration Library Case #02: Intra-subject Brain MR FLAIR to MR T1
Input
|
|
|
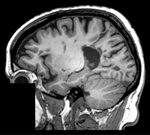
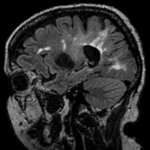
| fixed image/target | moving image | |
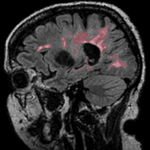
| resampled labelmap in reference space |

|
|
| result | resample | segmentation labelmap |
Modules
- Slicer 3.6.1 recommended modules: BrainsFit, Robust Multiresolution Affine
Objective / Background
This scenario occurs in many forms whenever we wish to align all the series from a single MRI exam/session into a common space. Alignment is necessary because the subject likely has moved in between series. As additional files we have a labelmap for the moving image we need to move along .
Keywords
MRI, brain, head, intra-subject, FLAIR, T1, defacing, masking, labelmap, segmentation
Input Data
- reference/fixed : T1 SPGR , 1x1x1 mm voxel size, 256 x 256 x 146, sagittal,
- moving: T2 FLAIR 1.2x1.2x1.2 mm voxel size, sagittal
- tag: segmentation labelmap obtained from above FLAIR, to be resampled with result transform
Registration Challenges
- the amount of misalignment is small. Subject did not leave the scanner in between the two acquisitions, but we have some head movement.
- we know the underlying structure/anatomy did not change, but the two distinct acquisition types may contain different amounts of distortion
- the T1 high-resolution had a "defacing" applied, i.e. part of the image containing facial features was removed to ensure anonymity. The FLAIR is lower resolution and contrast and did not need this. The sharp edges and missing information in part of the image may cause problems.
- we have one or more label-maps attached to the moving image that we also want to align.
- the different series have different dimensions, voxel size and field of view. Hence the choice of which image to choose as the reference becomes important. The additional image data present in one image but not the other may distract the algorithm and require masking.
- hi-resolution datasets may have defacing applied to one or both sets, and the defacing-masks may not be available
- the different series have different contrast. The T1 contains good contrast between white (WM) and gray matter (GM) , and pathology appears as hypointense. The FLAIR on the other hand shows barely any WM/GM contrast and the pathology appears very dominantly as hyperintense.
Key Strategies
- Slicer 3.6 recommended modules: BrainsFit, Robust Multiresolution Affine
- we use an affine transform with 12 DOF (rather than a rigid one) to address distortion differences between the two protocols
- we choose the SPGR as the anatomical reference. Unless there are overriding reasons, always use the highest resolution image as your fixed/reference, to avoid loosing data through the registration.
- the defacing of the SPGR image introduces sharp edges that can distract the registration algorithm; we use a multi-resolution approach (initialization + 6 DOF for BrainsFit or the Robust Multires module) to avoid instability
- because of the contrast differences and the defacing we use Mutual Information as the cost function (default for both recommended modules)
Procedures
with BrainsFit (ca. 3 min):
- download example dataset
- load into 3DSlicer 3.6
- open Registration : BrainsFit module
- Input Parameters: set SPGR as fixed and FLAIR as moving image
- Registration Phases:
- select Initialize with CenterofHeadAlign
- select Include Rigid registration phase
- select "Include Affine registration phase"
- Output Settings: select "New Linear Transform" under Output Transform
- accept all other defaults & click apply
- program will automatically move FLAIR image under the result transform; also manually move the labelmap
- right click on either image and select Harden Transform to apply & resample
- save result images/scene
with MultiresolutionAffine (ca. 15 min):
- download example dataset
- load into 3DSlicer 3.6
- open Registration : RobustAffineMultiresolution module
- set SPGR as fixed and FLAIR as moving image
- accept all defaults & click apply
- go to Data module and move FLAIR and labelmap under the result transform
- right click on either image and select Harden Transform to apply & resample
- save result images/scene
for more details see the tutorial under Downloads
Registration Results
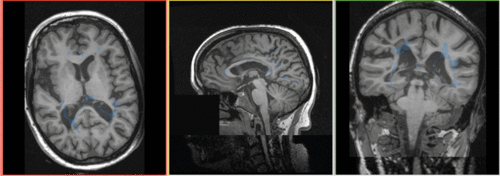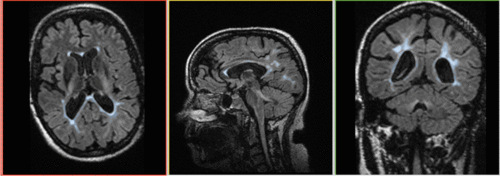 unregistered
unregistered
Robust Multiresolution Registration Result: FLAIR + segmentation aligned with SPGR : registered w. Multiresolution affine
BrainsFit Result: FLAIR aligned with SPGR: registered w. BrainsFit
Conclusion: Both Multiresolution and BrainsFit modules produce good registrations without the need for additional initialization or masking. The BrainsFit solution is the faster of the two (3 minutes vs. 15 minutes).
Download
- DATA:Registration Library Case 02: MSBrain intra-subject multi-contrast (Data & Solution Xforms, zip file 18 MB)
- PRESETS: both recommended modules run with default settings. No presets required.
- TUTORIALS: