Difference between revisions of "Projects:RegistrationLibrary:RegLib C37"
From NAMIC Wiki
| Line 43: | Line 43: | ||
###select a new transform "Linear Transform", rename to "Xf1_affine" | ###select a new transform "Linear Transform", rename to "Xf1_affine" | ||
##Leave all other settings at default | ##Leave all other settings at default | ||
| − | *'''Phase | + | *'''Phase II: [optional]: generate skull mask |
| − | ###select a new volume "Output Image Volume'', rename to "DTI_base_Xf1" | + | #open ''Skull Stripper for Structural MRI'' Module (Extension module) |
| + | #increase ''Iterations" to 200 and ''Subdivisions'' to 12 | ||
| + | #both results will have to be edited manually using the Editor module. | ||
| + | *'''Phase III: Register BSpline''' | ||
| + | #open Registration : ''BrainsFit'' module (presets: BRAINSfit_Xf3 or _Xf4 | ||
| + | ##select a new volume "Output Image Volume'', rename to "DTI_base_Xf1" | ||
##Registration Parameters: increase ''Number Of Samples'' to 200,000 | ##Registration Parameters: increase ''Number Of Samples'' to 200,000 | ||
##Registration Parameters: set ''Number Of Grid Subdivisions'' to 7x7x7 | ##Registration Parameters: set ''Number Of Grid Subdivisions'' to 7x7x7 | ||
##Leave all other settings at default | ##Leave all other settings at default | ||
| + | ##if using masks, set the ''Mask Processing Mode'' to "ROI" and select the two labelmaps generated in phase II as Input Masks | ||
=== Registration Results=== | === Registration Results=== | ||
Revision as of 21:19, 3 March 2011
Home < Projects:RegistrationLibrary:RegLib C37Back to ARRA main page
Back to Registration main page
Back to Registration Case Library
Contents
v3.6.1  Slicer Registration Library Case #37:
Slicer Registration Library Case #37:
Intra-subject Brain MRI: T1 Tumor Growth / Resection Assessment
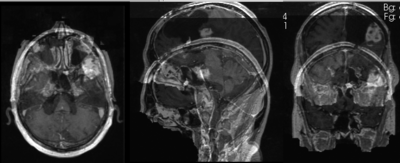
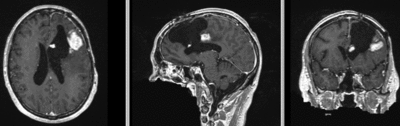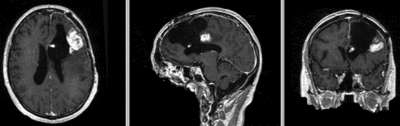
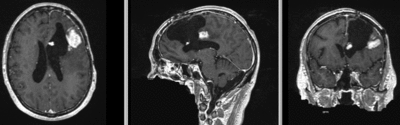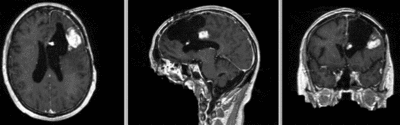
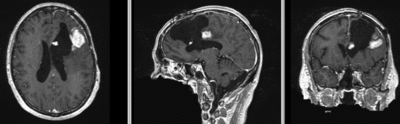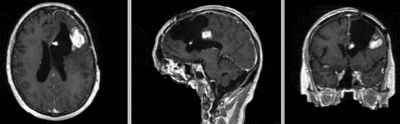
Input
|
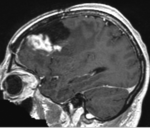
|
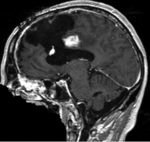
|
| fixed image/target | moving image |
Modules
Slicer 3.6.1 recommended modules:
Objective / Background
This is a typical case of change assessment.
Keywords
MRI, brain, head, intra-subject, T1, tumor, change assessment
Input Data
- reference/fixed : T1 SPGR , 1x1x1 mm voxel size, 256x256x176 ,sagittal
- moving: T1 SPGR , 1x1x1 mm voxel size, 256x256x176 ,sagittal
Overall Strategy
- run rigid/affine registration (BRAINSfit) with "CenterOfHeadAlign" initialization
- run 7x7x7 nonrigid regista=ration (BRAINSfit BSpline) using the above affine as input
- alternative: run skull stripper module on both images to generate masks for the nonrigid registration
Procedures
- Phase I: Register Rigid/Affine
- open Registration : BrainsFit module (presets: BRAINSfit_Xf1 or _Xf2
- Registration Phases:
- select/check Include Affine registration phase
- for affine also select/check Include Scale Versor 3D and Include Affine registration phase
- Registration Parameters: increase Number Of Samples to 200,000
- select a new transform "Linear Transform", rename to "Xf1_affine"
- Leave all other settings at default
- Registration Phases:
- Phase II: [optional]: generate skull mask
- open Skull Stripper for Structural MRI Module (Extension module)
- increase Iterations" to 200 and Subdivisions to 12
- both results will have to be edited manually using the Editor module.
- Phase III: Register BSpline
- open Registration : BrainsFit module (presets: BRAINSfit_Xf3 or _Xf4
- select a new volume "Output Image Volume, rename to "DTI_base_Xf1"
- Registration Parameters: increase Number Of Samples to 200,000
- Registration Parameters: set Number Of Grid Subdivisions to 7x7x7
- Leave all other settings at default
- if using masks, set the Mask Processing Mode to "ROI" and select the two labelmaps generated in phase II as Input Masks
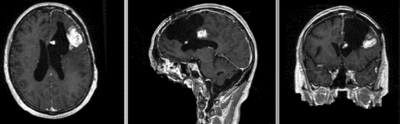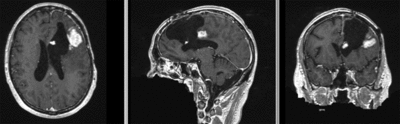
Registration Results
| original unregistered | |
| rigid registered | affine registered |
| nonrigid registered (7x7x7 BSpline unmasked) | nonrigid registered (7x7x7 BSpline masked) |
Download
- Data
- Presets