Projects:RegistrationLibrary:RegLib C39
From NAMIC Wiki
Home < Projects:RegistrationLibrary:RegLib C39
v3.6.1
Back to ARRA main page
Back to Registration main page
Back to Registration Use-case Inventory
Contents
v3.6.1  Slicer Registration Library Case 39: In Vitro cerebral body CT (thoracic spine)
Slicer Registration Library Case 39: In Vitro cerebral body CT (thoracic spine)
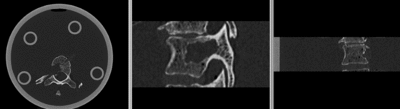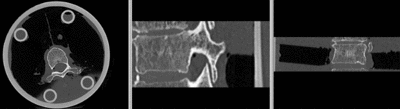
Input
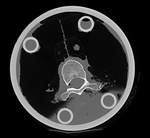
|
|

|
| baseline image | follow-up |
Modules
- Slicer 3.6.1 recommended modules: BRAINSFit
Objective / Background
This dataset contains in vitro CT of two human vertibral body segments (T10) before and after regional damage. Goal is to align the two images to study in detail the local changes that occurred.
Keywords
CT, spine cancer, vertebrae
Issues Challenges
- two images have very different spatial origins (as defined in original DICOM header): recenter first
- the baseline container contains the vertebra upside down (as apparent from orientation of the pedicle/roots of the vertibral arches coming out of the body, visible in sagittal or coronal view). Hence this image needs to have its z-axis (IS) reversed before registration
- the containers walls and lid contain dominant image content that would prevent a successful automated registration; cropping is required
- the two vertebrae have strong rotational misalignment; manual initial alignment is recommended
Input Data
- fixed: CT , 0.6 x 0.6 x 0.40 mm voxel size, axial; 292 x 292 x 91 unsigned short image
- moving: CT , 0.6 x 0.6 x 0.40 mm voxel size, axial; 512 x 512 x 107 unsigned short image
Procedure / Pipeline
- Open case scene file or import image data: RegLib_C28_SlicerScene1.mrml
- Place T10intact and T10defect in back- and foreground respectively.
- Scroll through sagittal view and note the inverted axis orientation
- Flip axis
- Go to the Data module
- right-click on the Scene node and select "Insert Transform Node" from the menu
- select the newly created transform node, then in the MRML Node Inspector below, rename it to "Xf0_AxialFlip"
- select the "T10intact" node and drag it inside/on top of the "Xf0_AxialFlip" node
- ensure you have T10intact selected for background and the fade slider set on background (B) so you can see changes happening.
- Go to the Transforms module
- Select "Xf0_AxialFlip" from the Transform Node menu
- In the 4x4 matrix displayed, click in the field containing the "1.0000" in row 3 and column 3.
- hit Return key to activate editing
- change the number to -1. Hit Enter.
- you should see the image flip in the sagittal and coronal slice views
- Go to the Data module
- Crop Volumes
- Go to the Crop Volume module
- select "T10intact" as Input Volume; select "Create new MRML ROI Node" in the ROI menu, select "create new volume" in the Output Volume" menu
- you should see a small blue rectangle in all 3 views. Click in the views to expand the region to contain only the L4 vertebra (body and processes)
- when finished, click on Do ROI resample
- go to back to the Data module and change the name of the new "SubVolume" to "T10intact_crop"
- delete the ROI nodes (right click: delete node)
- repeat the same for the "T10defect" image
- Registration
- Go to the BRAINSfit module
- 'Presets: select the "BRAINSFit_Affine" from the parameter set menu. See here for a guide on how to Load/Save Registration Parameter Presets]]
- otherwise select the following parameters:
- Fixed Image Volume: :T10intact_crop ; Moving: T10defect_crop
- check boxes for "Moments align", Include Rigd registr. phase , Include ScaleVersor3D, include Affine
- Slicer Linear Transform: select "create new transform", rename to "Xf1_Affine" or similar
- leave rest at defaults. Click Apply
- registration should take ~ 10 secs.
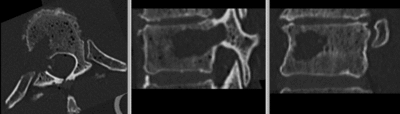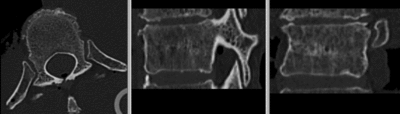
- use fade slider to verify alignment; compare with result snapshots shown below.
- Go to the BRAINSfit module
Registration Results
Download
- Data
- Presets