NA-MIC External Collaborations
Back to NA-MIC Collaborations
Contents
Projects funded by "Collaborations with NCBC PAR"
This section describes external collaborations with NA-MIC that are funded by NIH under the "Collaboration with NCBC" PAR. (Details for this funding mechanism are provided here).
|
PAR-08-183: R21EB009900 Johns Hopkins Skull Stripping The group at Johns Hopkins is developing software that enables the stripping of skull, scalp, and meninges from structural MRI scans in a fully automated fashion. Funding Duration: 08/01/2009-07/31/2011 |
|
PAR-07-249: R01AA016748 Measuring Alcohol and Stress Interaction with Structural and Perfusion MRI This project is funded under an NCBC collaboration grant to PIs James Daunais, Robert Kraft, and Chris Wyatt. The goal of this project is to examine the the effects of chronic alcohol self-administration on brain structure and function the monkey brain. MRI image analysis tools from the NA-MIC kit will be adapted for use with the monkey brain datasets. More... Funding Duration: 07/15/2009-03/31/2010 |
|
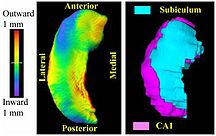
PAR-07-249: R01EB008171 3D Shape Analysis for Computational Anatomy This project is funded under an NCBC collaboration grant to PI Michael Miller JHU (with Joe Hennessey) Funding Duration: 05/21/2009-02/28/2013 |
|
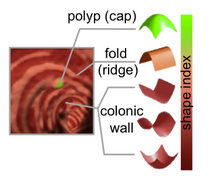
PAR-07-249: R01CA131718 NA-MIC Virtual Colonoscopy This project is funded under an NCBC collaboration grant to PI Hiroyuki Yoshida. The goal of this project is to More... Funding Duration: 01/26/2009-11/30/2012 |
|
PAR-07-249: R01MH084795 The Microstructural Basis of Abnormal Connectivity in Autism This project is funded under an NCBC collaboration grant to PI Janet Lainhart, MD. It will use tools developed within NAMIC for a longitudinal neuroimaging, clinical, and neuropsychological study of late neurodevelopment in autism.combining analysis of connectivity and morphometry. Funding Duration: 01/04/2009-01/31/2014 |
|
PAR-07-249: R01EB006733 Development and Dissemination of Robust Brain MRI Measurement Tools This project is funded under an NCBC collaboration grant to PI Dinggang Shen at UNC-Chapel Hill. The goal of this project is to develop and widely distribute a software package for robust measurement of brain structures in MR images using computational neuroanatomy methods.More... Funding Duration: 09/17/2008-08/31/2011 |
|
PAR-05-063: R01CA124377 An Integrated System for Image-Guided Radiofrequency Ablation of Liver Tumors This project is funded under an NCBC collaboration grant to PI Kevin Cleary at Georgetown University. The goal of this project is to develop and validate an integrated system based on open source software for improved visualization and probe placement during radiofrequency ablation (RFA) of liver tumors.More... Funding Duration: 09/14/2007-07/31/2012 |
|
PAR-05-063: R01EB005973 Automated FE Mesh Development This project is funded under an NCBC collaboration grant to PIs Nicole Grosland and Vincent Magnotta at UIowa. The goal of this project is to integrate and expand methods to automate the development of specimen- / patient-specific finite element (FE) models into the NA-MIC kit. More... Funding Duration: 09/20/2006-06/30/2011 |
Additional External Collaborations
This section describes external collaborations with NA-MIC that are funded by other mechanisms:
|
PAR-05-057: R01NS050568 BRAINS Morphology and Image Analysis This project is a funded under a Continued Development and Maintenance of Software grant to PIs Vincent Magnotta, Hans Johnson, Jeremy Bockholt, and Nancy Andreasen at the University of Iowa. The goal of this project is to update the BRAINS image analysis software developed at the University of Iowa. More... |
|
Children's Pediatric Cardiology Collaboration with SCI/SPL/Northeastern Collaboration with John Triedman, Matt Jolley, Dana Brooks, SCI. |
| U54EB005149-04S1 NA-MIC Collaboration with NITRC The NA-MIC Project is working to make NA-MIC neuroimaging software available through the NITRC web site. Supplemental support is helping to create the Slicer3 Loadable Modules project so that slicer plugins can be hosted on NITRC, allowing greater scalability for developers and users of Slicer. | |
|
P41RR013218 NA-MIC Collaboration with NAC NAC, the neuroimage analysis center, is a national resource center. NAC is relying on the NA-MIC kit for its general software environment. The mission of NAC is to develop novel concepts for the analysis of images of the brain and develop and disseminate tools based on those concepts. Several ARRA-funded supplements to the NAC grant have close ties to related efforts in NA-MIC. |
|
U41RR019703 NA-MIC Collaboration with NCIGT NCIGT is leveraging the NA-MIC kit as a platform for developing dedicated IGT capabilities. |

|
R01CA111288 NA-MIC Collaboration with Prostate BRP BRP is leveraging the NA-MIC kit as a platform for developing dedicated IGT capabilities. |
|
Real-Time Computing for Image Guided Neurosurgery Using the Tera Grid to implement mesh-based non-rigid registration for Neurosurgery. |

|
UL1RR025758 NA-MIC support for Harvard CTSC Translational Imaging Consortium The Harvard CTSC Translational Imaging Consortium is using NA-MIC communication tools to facilitate the rapid deployment of expertise in medical imaging acquisition, analysis and visualization to clinical translational investigators. |
|
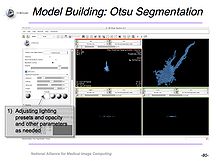
NCBC Supplement for Microscopy and Slicer An NCBC Supplement to NCMIR, UCSD focused on the utilization of Slicer with microscopy data and resulted in a tutorial for use of Slicer with confocal microscopy data. |
|
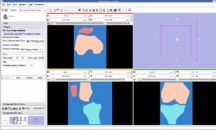
U54GM072970 NCBC Stanford Simbios Our sister NCBC at Stanford, dedicated to biomedical simulation, is working to adapt NA-MIC image analysis routines generate simulation models directly from MRI scans. |

|
U54LM008748 NCBC I2B2 Our sister NCBC at Harvard Medical School, dedicated to biomedical image informatics, is working with us through the Harvard based, NCRR funded CTSC (Catalyst) program, to develop common open source software for the community. |
|
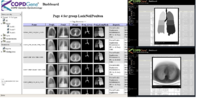
NAMIC supports COPDGene® quantitative analysis The Genetic Epidemiology of COPD (COPDGene®) Study is one of the largest studies ever to investigate the underlying genetic factors of Chronic Obstructive Pulmonary Disease or COPD. Through the enrollment of over 10,000 individuals, the COPDGene® Study aims to find inherited or genetic factors that make some people more likely than others to develop COPD. With the use of CT scans, COPDGene® also seeks to better classify COPD and understand how the disease may differ from person to person. |

|
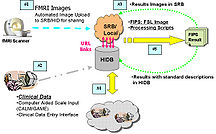
U24RR025736 BIRN CC The NCRR-funded efforts of the BIRN Coordinating Center (BIRN-CC) strive to apply state-of-the-art computer science techniques to the growing problem of working with large biomedical informatics datasets. Through a series of test bed projects and use case studies, the BIRN-CC refines and optimizes its offerings. NA-MIC provides an opportunity for BIRN-CC to work closely with the image analysis community and learn from the experience of NA-MIC DBPs. |
|
U24RR021992 fBIRN The Function BIRN (fBIRN) addresses the difficult problem of collecting and analyzing fMRI data collected at multiple sites in the context of schizophrenia research. A number of important and difficult scientific and engineering problems can only be addressed in the context of a multi-site consortium that aims to reproducibly quantify brain activity. Outcomes of fBIRN include quality assurance checks that have become industry standards, novel statistical approaches to identify and control for site-specific and scanner-specific biases, standardized stimulus paradigms, and fMRI informatics techniques for large scale datasets. |
|
U24RR021382 mBIRN The Morphometry BIRN (mBIRN) seeks to support multi-site brain studies using structural and diffusion MRI. Standard acquisition protocols, analysis methods, and informatics techniques have all been studied and promulgated by the mBIRN community of researchers. |

|
U24RR026057 Collaborative Tools Support Network for BIRN As the mBIRN and fBIRN test bed activities wind down the activities, research labs that have adopted the BIRN tool suite are supported through the BIRN-CTSN efforts. This funding covers software maintenance and ongoing deployment support to ensure that these important resources remain a vital part of the research community. |

|
BrainColor brainCOLOR is a Collaborative Open Labeling Online Resource for to create high quality manually segmented brain data sets. |
International Collaborations

|
Ontario Consortium of Adaptive Interventions for Radiation Oncology (OCAIRO) OCAIRO is a cross-Ontario initiative led by Dr. Jaffray, and will work towards developing adaptive radiation therapy--a new approach involving the creation of hardware, software, imaging and database systems to enable oncologists to adapt radiation to each individual patient and their response during the course of therapy. |

|
Computer Aided and Image Guidance Medical Interventions (CO-ME) CO-ME The National Centre of Competence in Research (NCCR) Co-Me is a network of leading clinics and engineering sites in Switzerland with strong links to industry and international partners. |

|
Common Toolkit (CTK) CTK is a multi-institution international collaboration to share software development resources for medical imaging applications. |

|
Real Time Computer Simulation of Human Soft Organ Deformation for Computer Assisted Surgery |
| |
NA-MIC Collaboration with Research and Development Project on Intelligent Surgical Instruments Intelligent Surgical Instruments Projects uses Open-source software engineering tools developed by NA-MIC, and leverage it to surgical robotics. |
|
Vascular Modeling Toolkit Collaboration Slicer as a platform for segmentation and geometric analysis of vascular segments and image-based computational fluid dynamics (CFD). More... Collaboration with Luca Antiga of the Mario Negri Institute. |