Projects:RegistrationLibrary:RegLib C07
From NAMIC Wiki
Revision as of 13:50, 19 August 2011 by Meier (talk | contribs) (Created page with 'Back to ARRA main page <br> Back to Registration main page <br> [[Projects:RegistrationDocumentation:UseCaseInv…')
Home < Projects:RegistrationLibrary:RegLib C07
v3.6.3
Back to ARRA main page
Back to Registration main page
Back to Registration Use-case Inventory
Contents
v3.6.3  Slicer Registration Library Case #07: Prostate MRI follow-up
Slicer Registration Library Case #07: Prostate MRI follow-up
Input
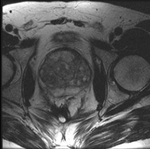
|
|
this is the time series of 2D images to be registered with the reference |
| fixed:prostate MRI day 1 |
moving:prostate MRI day 42 |
Modules
- Slicer 3.6.1 recommended modules: BrainsFit
Objective / Background
Evaluate changes
Keywords
MRI, prostate, follow-up
Input Data
- reference/fixed : axial MRI 256x256x36 ,0.7 x 0.7 x 4mm voxel size
- moving: axial MRI 256x256x20 ,0.8 x 0.8 x 5mm voxel size
Notes / Overall Strategy
- voxel size differs between the two images, also anisotropy is high: for the fine tuning a resampling to isotropic voxels for both volumes is recommended
- the large FOV includes a lot of additional image content that can be useful in constraining the alignment but also limits precision on the region of interest. Ev. cropping or masking to an ROI containing the prostate only is likely to improve the quality of the final registration
- the surface coils cause strong field inhomogeneity artifacts, apparent as strong intensity bias in both images. Bias correcion is recommended as preprocessing step.
- Overall strategy:
- Bias correction of both images
- Affine automated registration of Day42->Day1
- Low-level BSpline registration (5x5x3) on global image, using above affine as starting point
- Crop both images to prostate region only, resample to isotropic sizes
- and apply above registration to cropped version of Day42
- BSpline registration of cropped images only
- Reapply final transform to entire (uncropped image)
Procedures
- Phase I: Bias field correction
- load reference image and one moving image from the series
- open N4ITKBiasFieldCorrection module
- optional: select presets ("N4_Day1_axial" ) for module parameters from Parameter Set menu, or:
- select "Day01_axial" as input image, create new "Day1_axial_N4" output image
- set Iterations to 90,60,40
- set BSpline grid resolution to 2,2,1
- set Shrink factor to 3
- click Apply
- repeat above for the other image: "Day42_axial"
- Phase II: Affine Alignment
- go to BRAINSfit module
- Select Fixed Image = Day01_axial_n4 and Moving Image as "Day42_axial_n4"
- check boxes to include "rigid","scale", "skew" and "affine" registration phases
- select/check Include BSpline registration phase
- Output Settings:
- select a new transform "Slicer BSpline Transform", rename to "Xf1_moving_??"
- select a new volume "Output Image Volume, rename to "moving_??_Xf1"
- Registration Parameters: increase Number Of Samples to 200,000
- Registration Parameters: set Number Of Grid Subdivisions to 5,5,5
- Leave all other settings at default
- click: Apply; (runtime < 10 sec. on MacPro)
- adjust grid size until registration is acceptable
- you can see the commandline text of the registration performed by opening the Window/Error Log window and clicking on BRAINSfit commandline input
- Phase II: Batch Run
- open a terminal window
- via a TextEditor or prototyping/scripting software (e.g. Matlab), copy and modify the prototype line below, by changing only the moving input image:
/Applications/Slicer36/Slicer3 --launch /Applications/Slicer36/lib/Slicer3/Plugins/BRAINSFit --useBSpline --splineGridSize 5,5,5 --outputVolumePixelType short / --numberOfSamples 200000 --costMetric MMI --fixedVolume Reference/refLung_001.dcm --movingVolume Moving/Moving_001/Moving_001.dcm / --bsplineTransform Xforms/Moving_001_XfBSpl5.tfm --outputVolume MovingResampled/Moving_001_r.nrrd >> Logs/Moving_001_RegLog.txt 2>&1
- replace "/Applications/Slicer36" with your path of 3DSlicer
- create result directories MovingResampled, Logs, Xforms
- note that because input image is DICOM, and images are 2D only, each image of the time series must be in its own directory, otherwise Slicer will read them as a volume.
- paste all commands into a terminal window, or copy into a shell script and execute.
- Phase III: Evaluate Transform files
- upon completion, read the transform files with an editor and extract the displacements of interest
- The ITK transform files describe displacements at the grid nodes, many of which are outside the region of interest. Because BRAINSfit pads the grid with 1 voxel, the grid returned is actually 8x8x8. We use only the plane (*,*,4) for analysis and discard the y-direction displacements, which as expected are all zero.
- for details on the ITK transform format see the FAQ here
- To obtain displacements at arbitrary coordinates, interpolate the transform into a deformation field, e.g. using this module (details FAQ here):
/Applications/Slicer3.6.3/lib/Slicer3/Plugins/BSplineToDeformationField
Registration Results
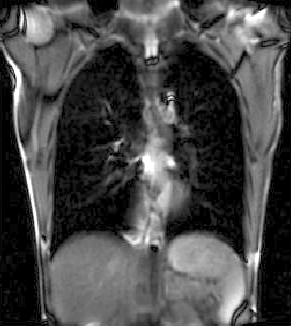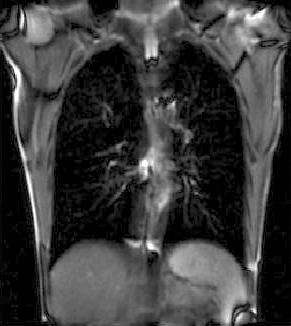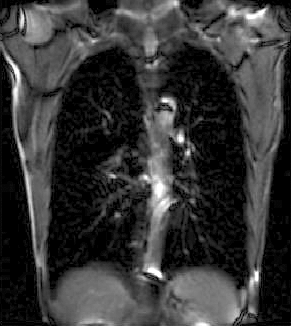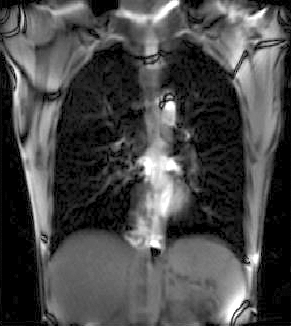
|
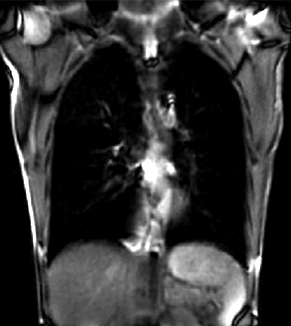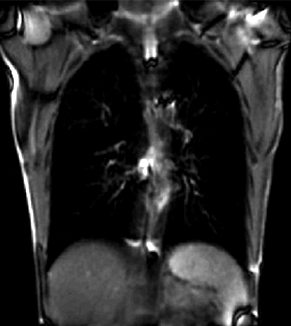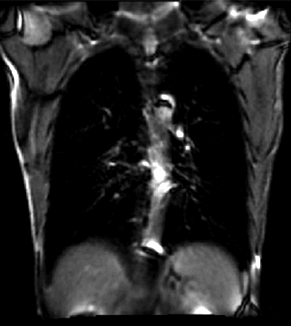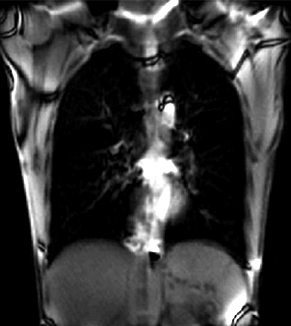
|
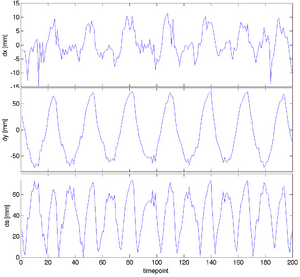
|
| unregistered moving series | moving input after registration (only frames 1-27 shown) | displacements (PDF) |
Download
- Data
- Results
Acknowledgments
Thanks to Amir Owrangi and Dr.Grace Parraga from the Robarts Institute for sharing the registration problem and data.