Difference between revisions of "2010 Winter Project Week SegmentationWizard"
(Added new images) |
|||
| Line 2: | Line 2: | ||
<gallery> | <gallery> | ||
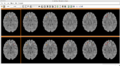
Image:PW-SLC2010.png|[[2010_Winter_Project_Week#Projects|Projects List]] | Image:PW-SLC2010.png|[[2010_Winter_Project_Week#Projects|Projects List]] | ||
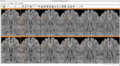
| − | Image: | + | Image:CompareViewFlairLesionDiffWholeBrain.png|Flair with lesion diff. |
| − | Image: | + | Image:CompareViewFlairLesionDiff.png|Lesions Time1 Vs. Time2 |
</gallery> | </gallery> | ||
Revision as of 22:00, 6 January 2010
Home < 2010 Winter Project Week SegmentationWizardKey Investigators
- MRN: Jeremy Bockholt, Mark Scully
- BWN: Steve Pieper
Objective
To implement a high level wizard that performs all necessary pre-processing steps and then segments lesion images.
Approach, Plan
1. Verify that all required sub-modules are production ready.
2. Diagram all possible branches with requirements.
3. Use the python scripting system to tie all the different modules together with branching logic.
Progress
Prior to the meeting:
1. Identified a number of modules to incorporate assuming they are production ready (SkullStripping, BiasCorrection, ProbabilisticBiasCorrection, RegisterImages, BrainsConstellation, etc).
2. Implemented a number of other modules that were needed but didn't yet exist (JointIntensityStandardization, LupusLesionSegmenter)
At the meeting:
1. Meet with individuals responsible for relevant modules and identify any issues.
2. Work with Steve Pieper to implement prototype wizard.
3. Get feedback on wizard prototype.
4. Prototype Qt-base Wizard with help from Julien and Jean-Christophe