Difference between revisions of "Projects:QIN:3D Slicer Annotation Image Markup:Use cases"
From NAMIC Wiki
| Line 20: | Line 20: | ||
* Preprocessing: none | * Preprocessing: none | ||
| | | | ||
| − | [[Image:QINUseCase_Stanford.png]] | + | [[Image:QINUseCase_Stanford.png|500px]] |
|} | |} | ||
| Line 34: | Line 34: | ||
* Preprocessing: none | * Preprocessing: none | ||
| | | | ||
| − | [[Image:QINUseCase_NCI.png]] | + | [[Image:QINUseCase_NCI.png|500px]] |
|} | |} | ||
| Line 50: | Line 50: | ||
* Preprocessing: registration, motion correction, resampling (not included in the provided dataset) | * Preprocessing: registration, motion correction, resampling (not included in the provided dataset) | ||
| | | | ||
| − | [[Image:QINUseCase_MGH.png]] | + | [[Image:QINUseCase_MGH.png|500px]] |
|} | |} | ||
| Line 65: | Line 65: | ||
* Tools used: Slicer, Syngo via, Pinnacle, in-house tools | * Tools used: Slicer, Syngo via, Pinnacle, in-house tools | ||
| | | | ||
| − | [[Image:QINUseCase_Iowa.png]] | + | [[Image:QINUseCase_Iowa.png|500px]] |
|} | |} | ||
| Line 79: | Line 79: | ||
* Tools used: in-house Matlab | * Tools used: in-house Matlab | ||
| | | | ||
| − | [[Image:QINUseCase_Moffitt.png]] | + | [[Image:QINUseCase_Moffitt.png|500px]] |
|} | |} | ||
| Line 91: | Line 91: | ||
* Tools used: RT-specific tools | * Tools used: RT-specific tools | ||
| | | | ||
| − | [[Image:QINUseCase_Maastro.png]] | + | [[Image:QINUseCase_Maastro.png|500px]] |
|} | |} | ||
| Line 104: | Line 104: | ||
* Preprocessing: registration of the DCE, DWI, T1w parameters to T2w MRI | * Preprocessing: registration of the DCE, DWI, T1w parameters to T2w MRI | ||
| | | | ||
| − | [[Image:QINUseCase_BWH.png]] | + | [[Image:QINUseCase_BWH.png|500px]] |
|} | |} | ||
Revision as of 20:33, 4 January 2012
Home < Projects:QIN:3D Slicer Annotation Image Markup:Use casesThe following is the list of the annotation/markup use cases provided by the QIN groups. Use cases were described using the following template: File:QINUseCaseTemplate.doc
For the use case to be considered complete, it must include:
- textual description, based on the form above
- screenshots of the annotations as they appear in the tool used by the site
- images being annotated
- markup/annotations
- description of how the markup/annotations can be reproduced
Contents
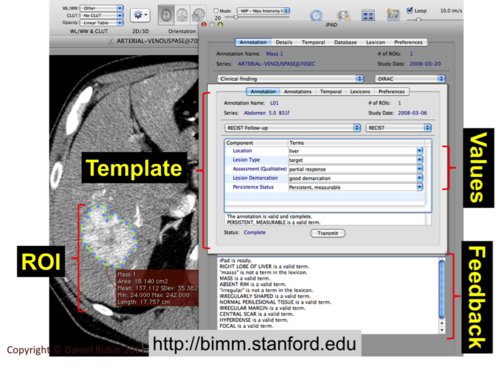
Stanford: Liver lesion
Use case incomplete, verbal description only
|
NCI: BreastDx
|
MGH: GBM
Use case partially complete, markup not included
|
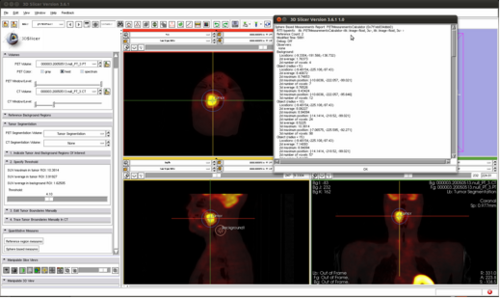
Iowa: Lung
Use case incomplete, verbal description only
|
Moffitt: Lung lesion
Use case incomplete, verbal description only. Masks and sample images are available (restricted access)
|
Maastro: Lung lesion
|
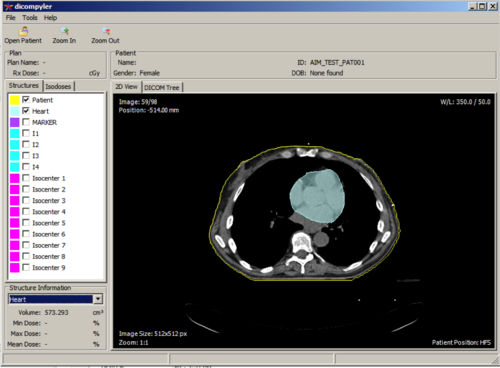
BWH: Prostate cancer
|