Difference between revisions of "Projects:RegistrationLibrary:RegLib C27"
From NAMIC Wiki
| Line 18: | Line 18: | ||
|- | |- | ||
|[[Image:Button_red_fixed.jpg|40px|lleft]] T2 FLAIR | |[[Image:Button_red_fixed.jpg|40px|lleft]] T2 FLAIR | ||
| − | |[[Image:Button_red_fixed.jpg|40px|lleft]] T1 | + | |[[Image:Button_red_fixed.jpg|40px|lleft]] T1 contr. |
| | | | ||
|[[Image:Button_green_moving.jpg|40px|lleft]] DTI Baseline | |[[Image:Button_green_moving.jpg|40px|lleft]] DTI Baseline | ||
Revision as of 14:42, 20 May 2010
Home < Projects:RegistrationLibrary:RegLib C27Back to ARRA main page
Back to Registration main page
Back to Registration Use-case Inventory
Contents
Slicer Registration Library Exampe #27: Diffusion Weighted Image Volume: align with structural reference MRI
Objective / Background
This is a typical example of DTI processing. Goal is to align the DTI image with a structural scan that provides accuracte anatomical reference. The DTI contains acquisition-related distortion and insufficient contrast to discern anatomical detail. For treatment planning and evaluation, location of functionally critical fiber tracts relative to the pathology is sought.
Keywords
MRI, brain, head, intra-subject, DTI, DWI
Input Data
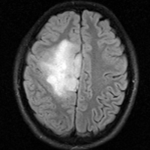
 reference/fixed : FLAIR axial, 0.4mm resolution in plane, 4mm slices
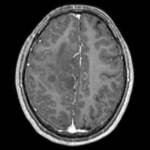
reference/fixed : FLAIR axial, 0.4mm resolution in plane, 4mm slices moving: Baseline image of acquired DTI volume, corresponds to T2w MRI , 1.96 x 1.96 x 3 mm voxel size, oblique
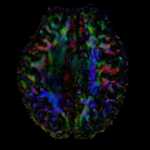
moving: Baseline image of acquired DTI volume, corresponds to T2w MRI , 1.96 x 1.96 x 3 mm voxel size, oblique tag: Tensor data of DTI volume, oblique, same orientation as Baseline image. The result Xform will be applied to this volume. The original DWI has 64 directions, the extracted DTI volume has 9 scalars, i.e. 128 x 128 x 40 x 9
tag: Tensor data of DTI volume, oblique, same orientation as Baseline image. The result Xform will be applied to this volume. The original DWI has 64 directions, the extracted DTI volume has 9 scalars, i.e. 128 x 128 x 40 x 9
Registration Results
Download
- download dataset (DTI estimate, baseline image FLAIR & T1) (NRRD files, zip file 15 MB)
- Presets
- Tutorial only
- Complete tutorial package
Link to User Guide: How to Load/Save Registration Parameter Presets
Discussion: Registration Challenges
- The DTI contains acquisition-related distortions (commonly EPI acquisitions) that can make automated registration difficult.
- the two images often have strong differences in voxel sizes and voxel anisotropy. If the orientation of the highest resolution is not the same in both images, finding a good match can be difficult.
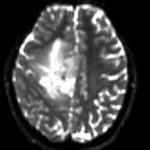
- there is widespread and extensive pathology in the right cortex that might affect the registration if its contrast is different in the baseline and structural reference scan.
Discussion: Key Strategies
- because the pathology appears similar in the FLAIR as in the DTI baseline, we choose the FLAIR as reference
- masking is likely necessary to obtain good results.
- in this example the initial alignment of the two scans is pretty good already. No initial affine alignment is needed.
- these two images are not too far apart initially, so we reduce the default of expected translational misalignment
- because speed is not that critical, we increase the sampling rate from the default 2% to 15%.