2010 Winter Project Week The Vascular Modeling Toolkit in 3D Slicer
Key Investigators
- Daniel Haehn, Student of Medical Informatics, University of Heidelberg, Germany
- Steve Pieper, Isomics, Inc.
- Luca Antiga, Mario Negri Institute, Italy
Objective
The Vascular Modeling Toolkit (VMTK) is a collection of libraries and tools for 3D reconstruction, geometric analysis, mesh generation and surface data analysis for image-based modeling of blood vessels. It should be very interesting to offer such techniques in 3D Slicer.
The official project page: http://www.vmtk.org/Main/VmtkIn3DSlicer
Approach, Plan
To provide VMTK functionality in 3D Slicer the vtkVmtk library has to be available as an add-on. With the connection of VMTK and 3D Slicer processing pipelines between VMTK code and other algorithms can be established.
Plan for project week: Clean-up of existing VMTK in 3D Slicer code, maybe adding additional functionality, investigating GWE possibilities..
Progress
The VMTK library and modules providing selected functionality are available as 3D Slicer extensions. This enables a flexible and convenient way for distribution and installation.
The following extensions are available:
- VmtkSlicerModule - a gui-less module providing the latest VMTK library (base installation)
- VMTKLevelSetSegmentation - providing the VMTK level-set segmentation process including different algorithms for initialization and evolution
- VMTKEasyLevelSetSegmentation - an easier interface to level-set segmentation with selected algorithms
- VMTKVesselEnhancement - Vesselness filtering to enhance tubular structures
- VMTKCenterlines - Centerline computation of polydata models
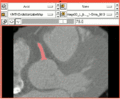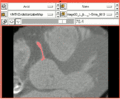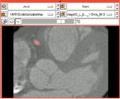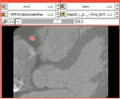
The modules have been successfully applied to segmentation problems (e.g. Coronary Artery Centerline Extraction - see Tutorial).
Project Week Results
- VMTK in 3D Slicer ready to use with Grid Wizard Enterprise (http://www.gridwizardenterprise.org)
- S2EL files to access the VMTK functionality. (Import of the logic classes)
- For example Frangi's Vesselness:
${SIGMA_MIN}=$range(0.1,1.0,0.1)
${SIGMA_MAX}=$range(1.0,4.0,0.5)
${SIGMA_STEPS}=$const(10)
${ALPHA}=$range(0.1,1.0,0.1)
${BETA}=$range(5.0,10.0,1.0)
${GAMMA}=$range(5.0,10.0,1.0)
${OUTPUT}=$const(/home/hype/gwe/results/frangi_out-${SIGMA_MIN}-${SIGMA_MAX}-N${SIGMA_STEPS}-${ALPHA}-${BETA}-${GAMMA}.nrrd)
${SLICER_HOME}/Slicer3 --no_splash --evalpython
"import sys;
from Slicer import slicer;
sys.path.append(str(slicer.Application.GetExtensionsInstallPath())+'/'+str(slicer.Application.GetSvnRevision())+'/VMTKVesselEnhancement/VMTKVesselEnhancement');
from SlicerVMTKVesselEnhancementGUI import *;
hiddengui = VMTKVesselEnhancement();
from SlicerVMTKVesselEnhancementLogic import *;
vesselness=SlicerVMTKVesselEnhancementLogic(hiddengui);
volNode=slicer.VolumesGUI.GetLogic().AddArchetypeVolume('/home/hype/gwe/liver.nrrd','Liver',0);
matrix = slicer.vtkMatrix4x4();
volNode.GetIJKToRASMatrix(matrix);
outVolumeData = vesselness.ApplyFrangiVesselness(volNode.GetImageData(),${SIGMA_MIN},${SIGMA_MAX},${SIGMA_STEPS},${ALPHA},${BETA},${GAMMA});
outputNode=slicer.MRMLScene.CreateNodeByClass('vtkMRMLScalarVolumeNode');
volumeNode=slicer.MRMLScene.AddNode(outputNode);
volumeNode.SetAndObserveImageData(outVolumeData);
volumeNode.SetIJKToRASMatrix(matrix);
volumeNode.SetModifiedSinceRead(1);
slicer.VolumesGUI.GetLogic().SaveArchetypeVolume(${OUTPUT},volumeNode);"
References
- Antiga L, Piccinelli M, Botti L, EneIordache B, Remuzzi A, Steinmann DA. (2008) An imagebased modeling framework for patientspecific computational hemodynamics. Med Biol Eng Comput 46(11):10971112
- Antiga L, Steinman DA (2008) The Vascular Modeling Toolkit. http://www.vmtk.org/
- Hähn D (2009) Integration of The Vascular Modeling Toolkit in 3D Slicer. Student Research Project, SPL.
- Piccinelli M, Veneziani A, Steinman DA, Remuzzi A, Antiga L (2009) A framework for geometric analysis of vascular structures: applications to cerebral aneurysms. IEEE Trans Med Imaging. In press.