DBP:Harvard:Software:Testing:DTI:Slicer
- DTMRI vs. DTMRI_Matlab:
- Table
- Updated table (12/13/05): Media:DTMRI_comparison.xls
- DTMRI vs LSDI (ADC/Trace):
- Table
- Updated table (12/13/05): Media:ADC.xls
- DTMRI vs. LSDI (FAI):
- Table
- Updated table (12/13/05): Media:FAI.xls
11/15/05 More comparison of LSDIrecon vs. DTMRI
- Table with new testing data here
- Notes:
- Using Matlab scripts to compute this data appears to be safe, except for:
- LSDIrecon DICOM files
- Marc: calculating Mode with Matlab?
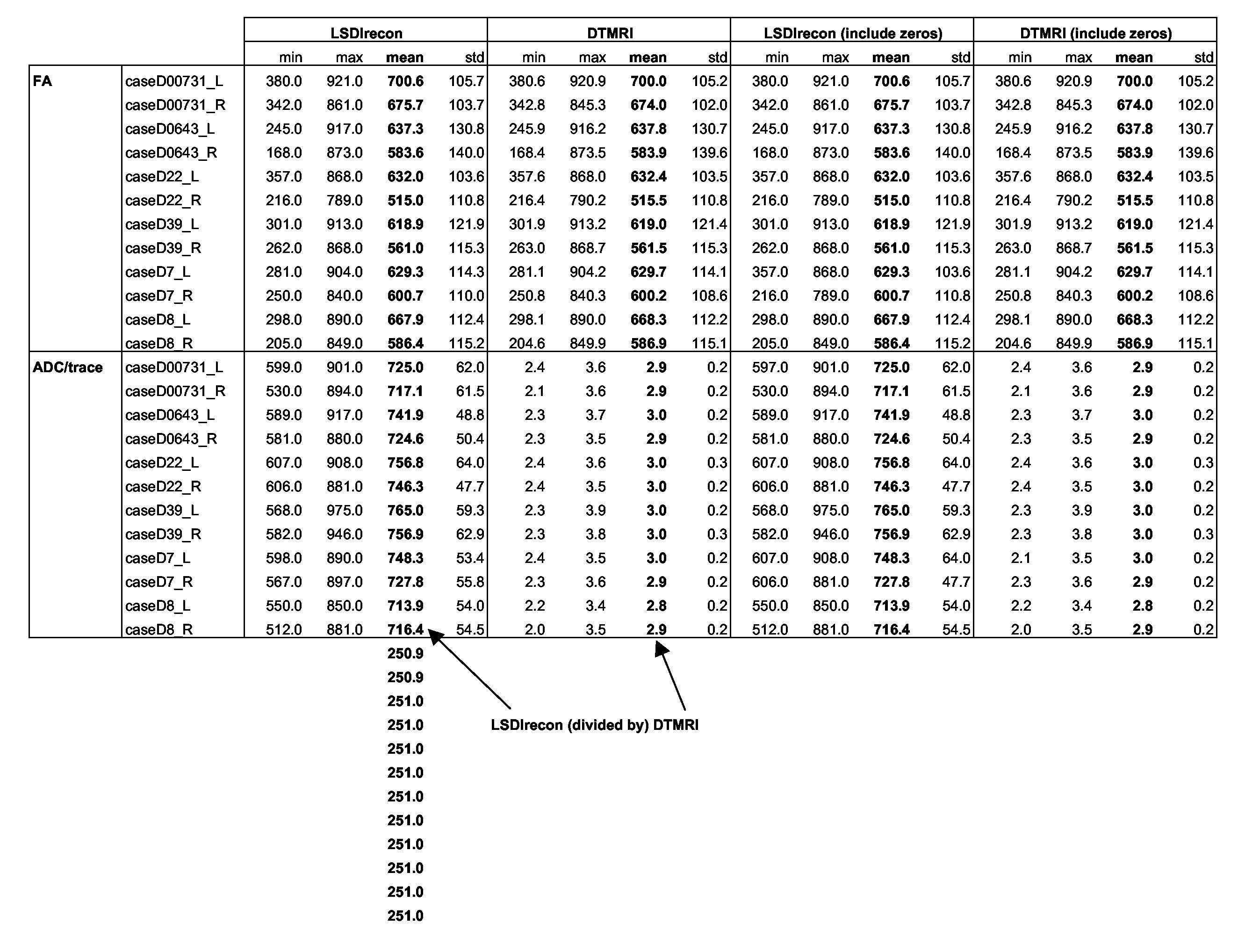
- ADC vs. Trace: Why the 251X difference?
- NRRD headers give same results as DTMRI
- Can we use Matlab scripts on DTMRI scalar measures for our studies?
- Would this mean not using LSDIrecon?
- Using Matlab scripts to compute this data appears to be safe, except for:
9/28/05 Comparison of LSDIrecon vs. DTMRI
- Trace values are very small compared to ADC (may be a Scale Factor issue)
- We have tried testing this using NRRD header data (as suggested), but we need Gordon to confirm that we are doing it correctly. Next week...
- The "Include Zeros" button in slicer_lmi DOES work correctly.
- FAI and trace values are NOT different when including zeros.
- Does this mean there are no actual "zero" values?
- Now that the button is there, do we want to keep including zeros?
- Currently the default is set to include zeros in slicer_lmi, but the function does not exist in slicer24, etc.
- FAI and trace values are NOT different when including zeros.
9/21/05 Comparison of LSDIrecon vs. DTMRI, using FA & ADC/trace as test variables. Also a comparison of the "include zeros" functionality that Steve included.
- Differences in FA are always <0.25% (usually much smaller)
- Raul removed the Scale Factor, which seems to make Trace values very small compared to ADC
- BUT DTMRI trace values are still consistent (in relative terms) with LSDIrecon ADC, i.e., always 250X smaller
- Including zeros (Steve's added button) did not make a difference. Does this mean 1) there is no difference, or 2) the "include zeros" button does not work?
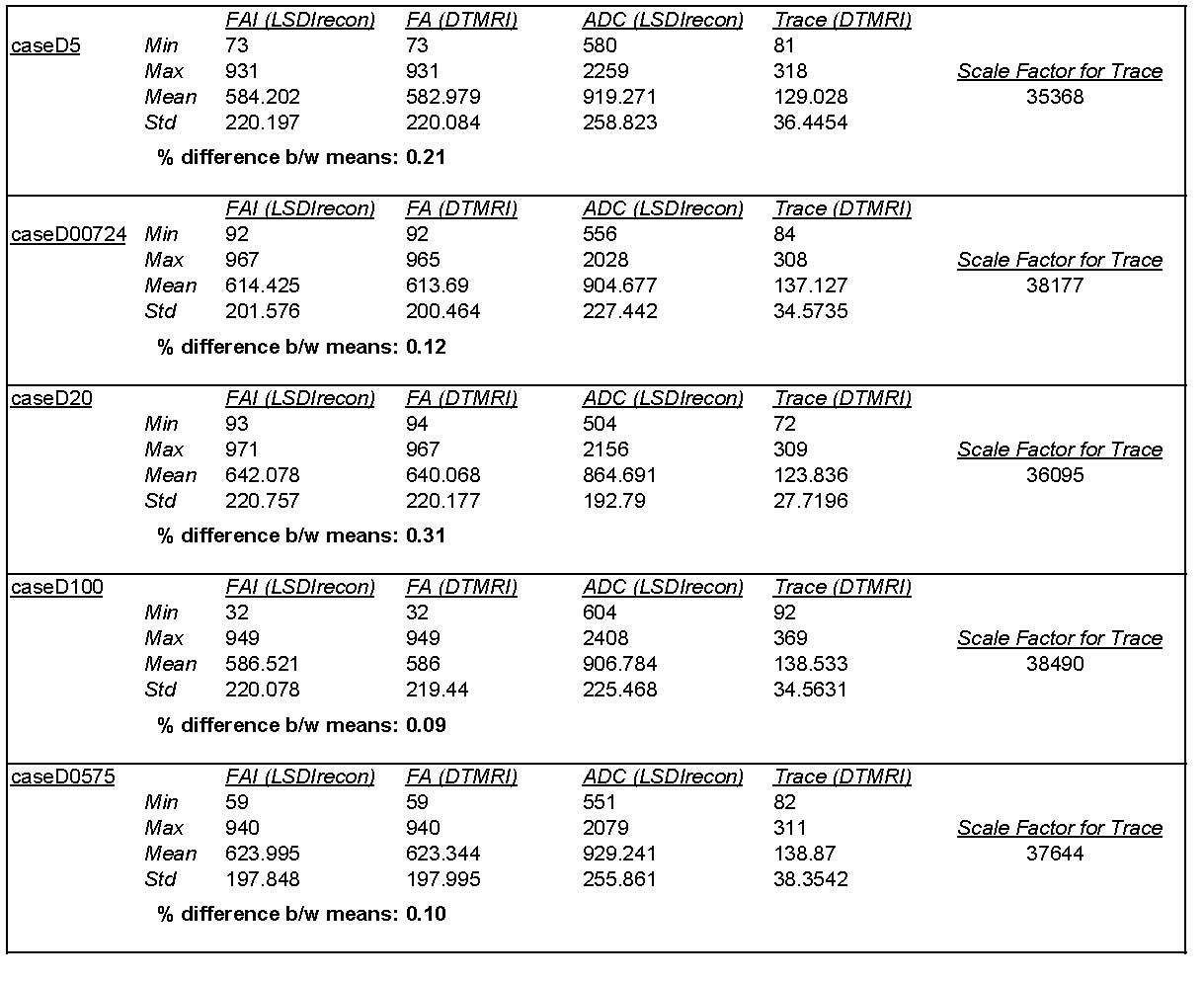
6/14/05: Comparison of LSDIrecon output (FA and Diffusivity) to DTMRI scalar volumes
- Differences in FA are small (typically less than 0.25%), but why are there differences?
- Differences in ADC/Trace exist, probably due to the Scale Factor. Mark has emailed Raul about this.
- Can other measures be compared between LSDIrecon and DTMRI?
- LSDIrecon outputs: AAI, ADC, DWI, FAI, FIL, L1x, L1y, L1z, RAI, T2W, VAI
- See a description of Stephan's LSDIrecon program (from /projects/schiz/software/bin/LSDIrecon.HOWTO, 6/15/05, but is this an older version of the file, 2003?)
- Note: Diffusion images were sagittally acquired, and the labelmap used was Mark's manual corpus callosum drawings
6/1/05: Sylvain and Mark met with Stephan a couple weeks ago to discuss the slice corruption problem with the lsdi2dwi script and DICOM cases (see notes from 5/13/05). Stephan discovered the source of the problem: the swab function behaves slightly differently under Linux and SUN.
- Stephan fixed the problem
- Sylvain implemented the code in the lsdi2dwi script
- Mark confirmed that the new script is reliable.
- It is now safe to use the lsdi2dwi-all.pl script on DICOM cases.
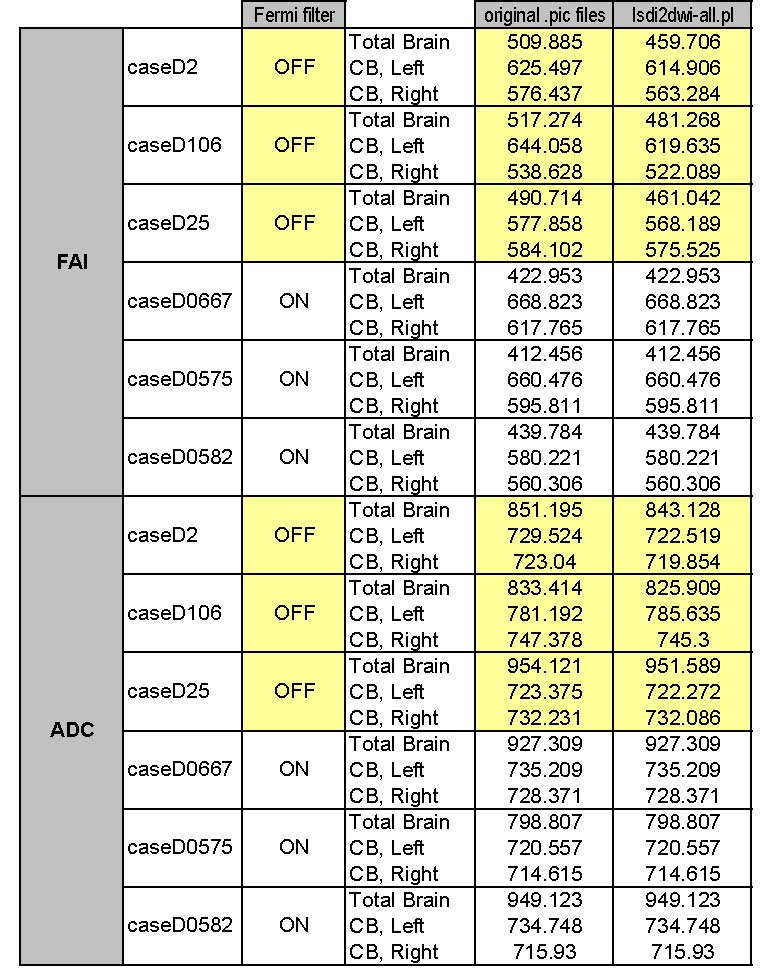
5/20/05: Below is the table confirming that Gordon's matlab scripts accurately differentiate Fermi-filtered from non-filtered cases.
- NOTE: T2W or DWI must be available to use this method.
Notes for this table:
- "Fermi filter" column was determined from Gordon's images, and confirmed using the test data below.
- Note that for highlighted cases, data in the "original .pic file" column was NOT filtered, and results are different from data using the Fermi-filtered lsdi2dwi script.
5/18/05: Gordon's results
5/17/05: Erin and Mark found out that a majority of cases were potentially reconstructed before Stephan's LSDIrecon v1.3 was implemented, so they are working with Gordon on a method to retroactively determine whether cases were reconstructed with a Fermi filter.
- Question for Stephan: What were the exact dates that changes in LSDIrecon were created?
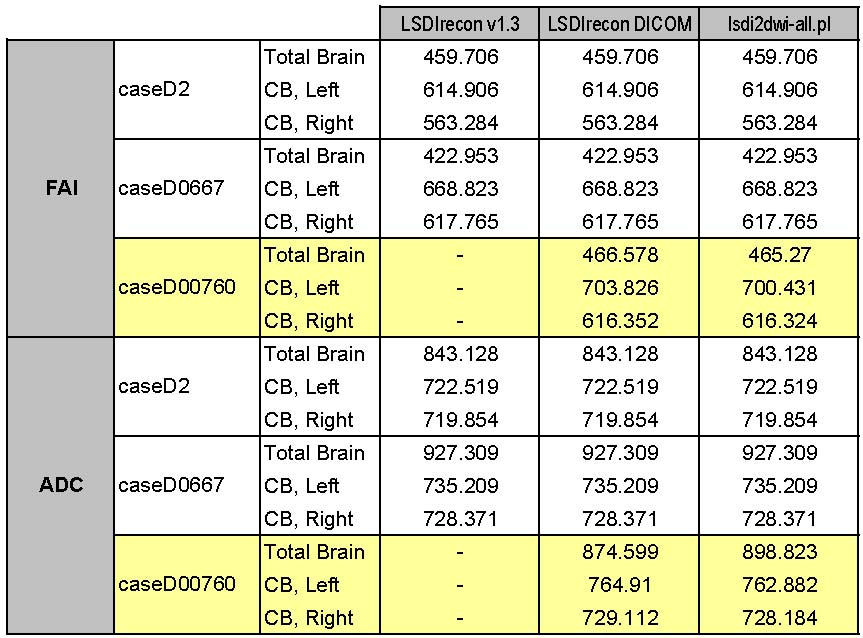
5/13/05: Sylvain modified the lsdi2dwi-all.pl script, and Erin & Mark compared the output of this script with the original versions of the LSDIrecon scripts. The output table is below.
Important notes/summary:
- The lsdi2dwi-all.pl script is (as of now) reliable for GENESIS CASES ONLY
- Two problems were discovered regarding the use of lsdi2dwi-all.pl script with DICOM cases:
- Output is in Little Endian format
- Several slices in each case become corrupt, leading to slight changes in FA/ADC values (see highlighted cases below)
- We will once again recruit help from Stephan to fix the problem of using this new script with DICOM cases.
- For now, it is ok to use the lsdi2dwi-all.pl script WITH GENESIS CASES ONLY. DO NOT ANALYZE DICOM CASES UNTIL FURTHER NOTICE!
In the table below:
- CB = Cingulum Bundle
- caseD00760 (highlighted) is DICOM
5/11/05: Mark and Marek met with Stephan Meier and identified one source of the differences found in the LSDIrecon scripts.
- Data in /projects/schiz/diff/chronic/males was obtained using either LSDIrecon version 1.2 or 1.3, which differ in the use of a Fermi filter in the latter version
- It is unclear which cases were run with and without the Fermi filter, and it will be difficult to determine this in most cases
- It has been proposed that anyone using data in /projects/schiz/diff/chronic/males in the past 2 years should re-analyze their data
5/11/05: Stephan also brought up the issue of a threshold used in his LSDIrecon scripts, which could also change numbers slightly.
The following is a copy of the parameters in the LSDIrecon_par file:
- 1.3 # DO NOT CHANGE (LSDIrecon version)
- 1 # parameter file: 0=long, 1=short for LSDI, 2=short for other
- 1 # Fermi filter in read direction: 0=off, 1=on
- 1 # correct missing lines: 0=no, 1=normal, 2=strong
- 2 # correct eddy current distortion: 0=no, 1=yes, 2=yes incl. NEX
- -10 # clipping threshold (<0 noise multiple, =0 none, >0 absolute)
- 1000 # b-value for reconstructed DWI, 0=maximum b-value
- 1 0.2 2.2 0.4 -0.3 0.5 0.2 # fit order (order: >0 constant amplitudes, <0 tensor amplitudes), min ADC1, max ADC1, step ADC1, min ADC2, max ADC2, step ADC2
- 1 # data type: 0=Signa 5.4; 1=5.6, 5.7, LX
- 1 # output: 0=nothing (for testing), 1=basic, 2=basic with header, 3=everything, 4=everything with header
- ./ # Path for output files, use ./ for local directory
- test # Prefix for output file names
- ./ # Path for input files, use ./ for local directory
- 0 # nr of images to offset during autoloading (usually 0)
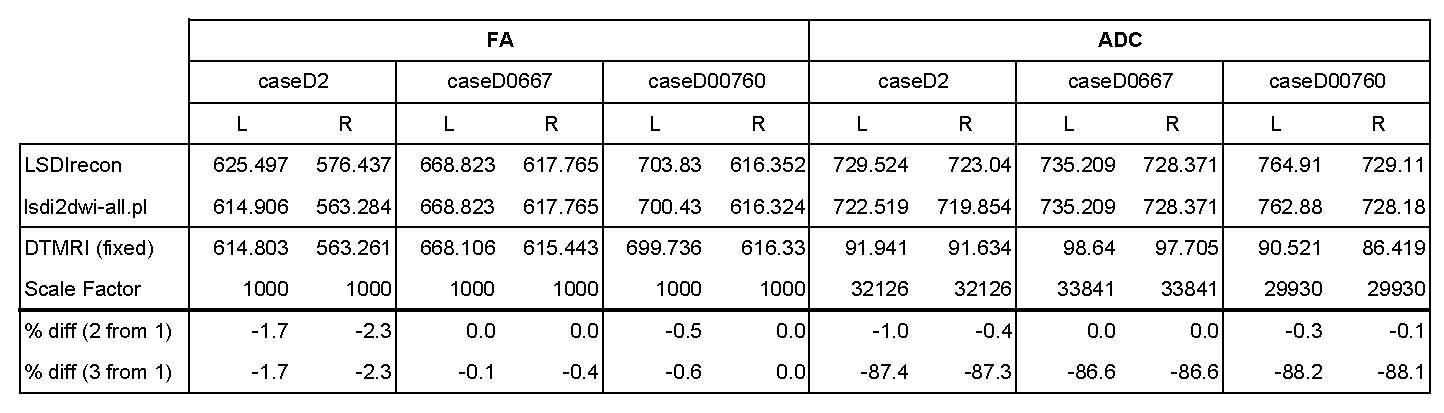
4/25/05: The DTMRI module in slicer_lmi now produces more reliable results (see below).
- A DICOM case is now included (caseD00760)
- DTMRI now gives results similar to the lsdi2dwi-all.pl script (but not exactly the same in all cases?)
- We must now determine how to use the "Scale Factor" to report correct ADC/Trace values
- The problem still exists of why the new lsdi2dwi-all.pl gives slightly different results from Stephan's old lsdirecon script (located in /projects/schiz/software/bin)
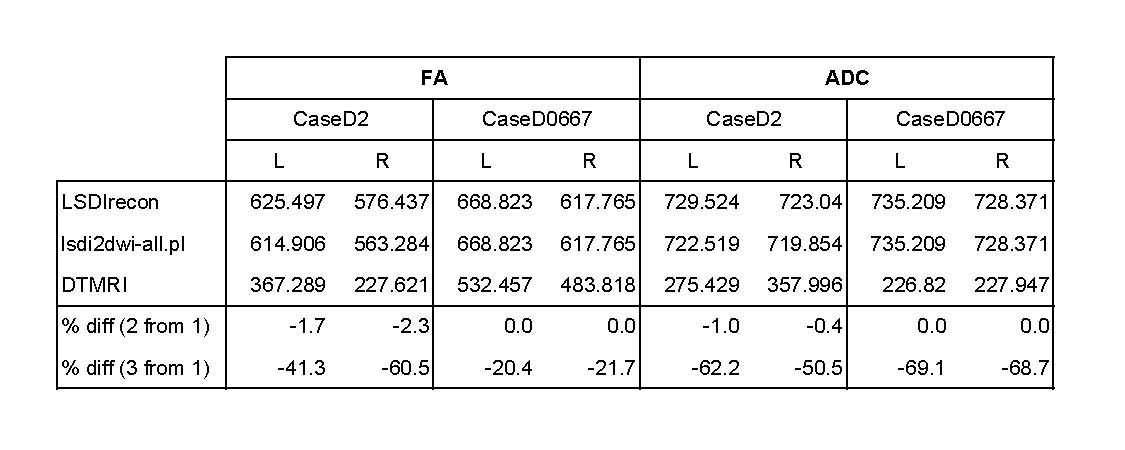
4/20/05: DTI data was obtained using 3 different "background" data sets:
- 1) Stephan's "old" LSDIrecon scripts
- 2) the new lsdi2dwi-all.pl script created by Sylvain and Stephan
- 3) tensor data, by converting DWI data and creating Scalar volumes, then casting float --> short to mask using labelmaps
Notes:
- labelmaps were of the cingulum bundle used in Marek's study
- both cases shown were obtained in Genesis format
- caseD2 acquired 1/9/00
- caseD0667 acquired 7/19/03